Abstract
Euthynnus (family Scombridae) is a genus of marine pelagic fish species with a worldwide distribution that comprises three allopatric species: E. alletteratus, E. affinis and E. lineatus. All of them targeted by artisanal and commercial fisheries. We analyzed 263 individuals from Atlantic and Pacific Oceans using two genetic markers, the mtDNA Control Region (350 bp) and nuclear calmodulin (341 bp). The results obtained challenge the phylogeny of this group. We found a deep genetic divergence, probably at species level, within E. alletteratus, between the North Atlantic-Mediterranean and the Tropical East Atlantic. This deep genetic divergence was tested with several species delimitation methods. This complete phylogeographic association between the North Atlantic and the Tropical East Atlantic support the hypothesis of two cryptic species. In addition, population genetic heterogeneity was detected between the North East Atlantic–Mediterranean and North West Atlantic regions. Our results indicate two scales of differentiation in what is currently considered a single population. Accordingly, for management purposes, the populations of E. alletteratus, should be divided into a minimum of three management units. On the other hand, the high level of differentiation found in E. alletteratus contrasts with the shallow genetic divergence of E. affinis and E. lineatus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the marine realm, and specifically for the pelagic species, the apparent absence of marine barriers, together with high migratory potential, large population sizes and pelagic planktonic stages, contribute to mitigates the genetic differentiation among regions (Palumbi, 1994). However, there is an increasing number of examples of genetic substructuring in pelagic marine fishes (Waples, 1998; Smith et al., 2015; Pascual et al., 2017).
Some of these species of pelagic fishes are heavily targeted by artisanal and commercial fisheries as well as for recreational activities. Thus, the accurate assessment of population genetic structure is essential for the management and conservation of both the species and their populations. Because of their meat quality and abundance, the species of the family Scombridae, which include tunas, mackerels and bonitos, are one of the most valuable commercial species (Ward, 1995). The majority of scombrids are pelagic fishes capable of long migrations (Collette & Nauen, 1983) with extremely large populations (Collette et al., 2011; Juan-Jordá et al., 2011, 2013; Laconcha et al., 2015; Waples et al., 2018) and large effective population sizes (Ne) (Ely et al., 2005; Qiu et al., 2013; Laconcha et al., 2015). All these factors contribute to make the analysis of their genetic population structure challenging. For instance, there is a considerable amount of literature about the biology and stock identification on major pelagic species such as the Atlantic Bluefin tuna [Thunnus thynnus (Linnaeus, 1758)] (Carlsson et al., 2004; Rooker et al., 2007; Díaz-Arce & Rodríguez-Ezpeleta, 2019). However, for other scombrids, particularly the small bodied size species, even basic aspects of their biology and genetic population structure are poorly understood, as is the case for the little tunny, Euthynnus alletteratus (Rafinesque, 1810). In spite of its relevance as a commercial resource for traditional and local fisheries along its distribution no management recommendation has been approved by ICCAT (International Commission for the Conservation of Atlantic Tunas) (ICCAT, 2020).
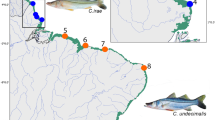
Members of genus Euthynnus are distributed worldwide in tropical and subtropical marine waters, and include three species distributed allopatrically: E. alletteratus displays an amphi-Atlantic and Mediterranean distribution, E. lineatus Kishinouye, 1920 is found primarily in the eastern Pacific Ocean, and E. affinis (Cantor, 1849) is found in the Indian and the central and western Pacific Oceans (Fig. 1) (Collette & Nauen, 1983). Distinction of these species is based on meristic characters and genetic markers. For example, E. alletteratus has no vomerine teeth but presents a higher gill raker count in the first arch than E. affinis and E. lineatus. The latter two, in turn, differ from each other on the total number of gill rakers, the total number of vertebrae and the presence or absence of vertebral protuberances (Collette & Nauen, 1983). Only two studies have used the mitochondrial marker COI to differentiate these three species (Kumar & Kunal, 2013; Santini et al., 2013). From a phylogenetic perspective, the genus Euthynnus was one of the latest groups that diversified within the scombrid family (Santini et al., 2013), thought to have appeared around 10 mya. Phylogenetic reconstructions and previous taxonomic studies agree that E. alletteratus would have diversified earlier from the common ancestor and that E. lineatus and E. affinis would be sister species that diverged more recently (Kumar & Kunal, 2013; Santini et al., 2013). Furthermore, although the genus Euthynnus has a worldwide distribution and occupies large areas where its members are subject to fisheries, there is scarce literature on the population genetics of its member species. No genetic evidences of population structure have been found in E. affinis among Indian locations (Kumar et al., 2012). Similarly, for E. alletteratus no genetic differentiation was detected within the Mediterranean (Allaya et al., 2015), nor between east and west Atlantic (Ward, 1995). However, preliminary results based on mtDNA (Alvarado Bremer & Ely, 1999) and on plastic and meristic characters (Gaykov & Bokhanov, 2008) suggests the presence of two stocks within the Atlantic Ocean.
Map depicting the distribution of the three recognized species of Euthynnus (Collette & Nauen, 1983). Sampling localities and the sample sizes at each location identified as follows: Portugal (PRT), Tunisia (TUN), Bermuda (BMU), Gulf of Mexico (GOM), Senegal (SEN), Côte d’Ivoire (CIV), Viet Nam (VNM), Mexico (MEX)
The aim of the present study is to analyse the population genetic structure of E. alletteratus and provide new information on phylogenetic relationships within genus Euthynnus. Since genetic markers have been proved to be an effective tool for species and stock identification, the results presented here can help in the elaboration of plans for stock assignment and improve the current management strategies for this species.
Material and methods
Dataset assembly
Samples of individuals representing the three recognized species of Euthynnus individuals were analysed for two unlinked genetic markers, the mitochondrial DNA control region (mtDNA CR) and the nuclear calmodulin intron 4 (CaM), based on previous protocols (Alvarado Bremer et al., 1996; Chow & Takeyama, 2000). The dataset for the mtDNA CR consisted of sequences of E. alletteratus (n = 175), E. affinis (n = 20) and E. lineatus (n = 10), collected by international observers and researchers on scientific surveys or in commercial fisheries from five different locations (see Table 1 and Fig. 1 for details). Specimens were initially assigned to species based on their geographical sampling localities, since they have allopatric and non-overlapping distribution (Collette & Nauen, 1983). No meristic counts nor morphological descriptions were recorded. Additionally, mtDNA CR dataset was complemented with 58 sequences of E. alletteratus obtained in previous studies (unpublished data, unless noted), from three locations, (see Table 1), totalling 263 sequences for this marker. CaM marker dataset was created with a selection of individuals of E. alletteratus (n = 30), E. affinis (n = 16) and E. lineatus (n = 10). Multiple sequence alignments were performed using ClustalW (Thompson et al., 1994) implemented in Geneious v7.1.9 (Kearse et al., 2012) and examined visually. CaM Sequences were phased using PHASE (Stephens et al., 2001) as implemented in DNAsp 6.12.03 (Rozas et al., 2017) in order to estimate the putative genotypes of the marker.
These sequence data have been submitted to the GenBank (www.ncbi.nlm.nih.gov/genbank) databases under accession number MN703152 - MN703207 and MN703208 - MN703342.
Genetic population analyses
The mtDNA CR haplotypes and CaM alleles were collapsed with DNAsp 6.12.03. Arlequin 3.5 (Excoffier & Lischer, 2010) was used to estimate the nucleotide (π) (Nei, 1987), haplotype diversity (h) (Nei & Tajima, 1981) indexes and expected heterozygosity (HS) as gene diversity, as well to perform hierarchical analyses of molecular variance (AMOVA) and the ϕST and FST pairwise comparisons (Wright, 1965; Excoffier et al., 1992). The comparison of samples from Côte d’Ivoire collected two decades apart (1998 and 2018) indicated temporal genetic homogeneity (ϕST = 0.006; P = 0.251 ± 0.004) in this region. Based on these results, haplotypes from these two samples were pooled together and used as a single location in the subsequent analysis.
mtDNA CR data was used to reconstruct demographic history of each genetically differentiated pool. Demographic histories were inferred with Bayesian Skyline Plots (BSP) performed in BEAST v1.10.4 (Drummond et al., 2012). Each model was elaborated using BEAUTi v1.10.4 (Drummond et al., 2012), to avoid misrepresentation of the data and overestimate the population size, the whole alignments instead of collapsed haplotypes were used for the analysis. HKY + G was selected as the best nucleotide substitution model (see below) and the mutation rate was fixed at 4.9 × 10−8 substitutions/site/year (Alvarado Bremer et al., 2005; Allaya et al., 2015). Chain length of 50 × 106, sampling every 1.000 generations, when needed, two runs were combined. Tracer v1.7.1 (Rambaut et al., 2018) was used to assess convergence with all ESS reaching the minimum values. Effective population size (Ne) was corrected by the generation time (τ) following the BEAST manual recommendation. Generation time was estimated using the approach of Theisen et al. (2008) adjusting to the life-history traits of E. alletteratus. Thus, considering age at first maturity and a life-span of seven years (Saber et al., 2018; Lucena-Frédou et al., 2021), we estimated the generation time in 3.25 years. Haplotype networks for the mtDNA CR were constructed with the Minimum Spanning method implemented in PopART (Leigh & Bryant, 2015).
Phylogenetic analyses and species delimitation
Due to the different size of the two datasets, the alignments were analysed separately without concatenation of the sequences. Two different approaches were used for the phylogenetic reconstructions of the respective datasets, maximum likelihood and Bayesian inference. The best nucleotide substitution model for each marker was inferred with MEGA 7 on the basis of the Bayesian Information Criterion (BIC) (Kumar et al., 2016). For mtDNA CR maximum likelihood reconstruction was performed in MEGA 7 (Kumar et al., 2016), using Tamura 3-parameter + Gamma (α = 0.208) as the best model with bootstrap 1,000 pseudoreplicates of node support. The sequences of three individuals belonging to the sister genus Auxis [Auxis rochei (Risso, 1810)] were used as outgroup. For CaM data maximum likelihood was performed using Jukes Cantor + Gamma + Invariable sites (I) (α = 0.05, I = 0.485) with 1,000 pseudoreplicates of bootstrap support.
Bayesian inference was also tested for both mtDNA CR and CaM data using BEAST v1.10.4 (Drummond et al., 2012) with two tree priors, namely Speciation Yule Process and Coalescent Constant Size (Yule, 1924; Gernhard, 2008). BEAUTi v1.10.4 (Drummond et al., 2012) was used to elaborate the different models for running BEAST. For mtDNA CR Yule and Coalescent tests were generated with the second-best nucleotide substitution model (HKY + G) since T92 was not available. The two tests were constructed under the assumption of strict clock and base frequencies estimated. The analysis was run with a MCMC (Markov Chain Monte Carlo) chain length of 10 million generations, sampling every 1.000 generations. For the CaM marker, Yule and Coalescent prior tests were constructed with Jukes Cantor + Gamma + Invariable sites, strict clock, base frequencies estimated and MCMC of 20 million generations sampling each 1.000 generations in both runs. In all runs, transition kernel states were checked to reach the minimum value and ESS were assessed to converge with Tracer v1.7.1 (Rambaut et al., 2018). Trees summary reconstructions were performed with TreeAnnotator v1.10.4 with a burn in of 10%, to avoid negative branches common ancestor heights option was selected. Final trees were represented graphically and edited with FigTree v1.4.3 (Rambaud, 2012). DA Genetic distances (Nei et al., 1983) among different phylogenetically differentiated groups were estimated in MEGA 7.
Different methods were used to identify putative species. For the ABGD (Automatic Barcode Gap Discovery) (Kekkonen & Hebert, 2014) three distances were tested, Jukes-Cantor (JC69), Kimura (K80) (TS/TV = 2) and Simple Distance. For mtDNA CR alignment parameters were set as follows Pmin: 0.001, Pmax: 0.1, Steps 10 and Relative Gap With: 1.5. while for CaM alignment settings were Pmin: 0.000001, Pmax 0.1, Steps 10 and Relative gap with 0.5. ABGD was run under the web server version available from https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html. MtDNA CR and CaM ultrametric trees generated under the two priors -Coalescence and Speciation- were used as input trees for GMYC (Generalized Mixed Yule Coalescent) (Pons et al., 2006), and single and multiple-threshold versions were tested https://species.h-its.org/gmyc/ (Zhang et al., 2013). Maximum likelihood trees were used for PTP (Poison Tree Process) and mPTP (multiPTP). These methods were run with the default options as implemented in the web server http://www.exelixis-lab.org/ (Kapli et al., 2017). For PTP and mPTP Likelihood Ratio Test (LRT) were calculated as − 2lnλ, being λ the relation between the null model likelihood and the likelihood of the model itself (Huelsenbeck & Bull, 1996). Finally, putative species were tested with bPTP (bayesian PTP) analyses (Zhang et al., 2013), for the two markers MCMC converge after 500.000 generations, thinning was set as 100 and a burn-in of 0.1, under the online version available from https://species.h-its.org/.
Results
The mtDNA CR sequence comparison included 103 polymorphic sites out of 350 bp, which defined 135 haplotypes (Table 1). CaM alignment revealed 10 polymorphic sites out of 341 bp, after allele phasing, the alignment resulted in 27 putative alleles.
Population genetics analyses
The highest values of haplotype diversity for the mtDNA CR were obtained in E. alletteratus from Bermuda and Gulf of Mexico, and in E. lineatus from Mexico’s East Pacific Coast. In each of those locations no haplotype was shared (Table 1). Conversely, the pooled samples of E. alletteratus from Portugal and Tunisia had the lowest haplotype diversity (h = 0.641 ± 0.061), with one haplotype shared in 46 out of 78 individuals (58.9%), while also displaying the lowest value of nucleotide diversity (π = 0.004 ± 0.003). Based on ϕST values of E. alletteratus in the North East Atlantic and Mediterranean area (NEA), no significant genetic divergence was found between the Portugal and Tunisia locations (ϕST = 0.007; P = 0.217 ± 0.004). A similar situation occurs in the North West Atlantic area (NWA) with no significant genetic differences between Bermuda and the Gulf of Mexico (ϕST = 0.054; P = 0.049 ± 0.002). Differentiation was also lacking < (ϕST = 0.011; P = 0.071 ± 0.007) among the two Tropical East Atlantic locations (TEA) Senegal and Côte d’Ivoire.
By contrast, a high degree of genetic heterogeneity between regions was observed (Table 2). High heterogeneity was detected between the NEA region and NWA region (ϕST = 0.405; P = 0.000 ± 0.000) (Table S1 in Supporting Information), although these regions shared some haplotypes. AMOVA showed that 37% of the variation corresponds to differences between regions (d.f 1; P = 0.000). In addition, significant high genetic differences were found between the North East Atlantic-Mediterranean region and Tropical East Atlantic region (TEA) (ϕST = 0.918; P = 0.000 ± 0.000) and also between the NWA and the TEA regions (ϕST = 0.866; P = 0.000 ± 0.000). Moreover, within the E. alletteratus none of the haplotypes was shared between the individuals from North Atlantic-Mediterranean (West and East) and the TEA area. Similarly, no haplotype was shared between the three Euthynnus species. The comparison between E. affinis (Viet Nam) versus E. lineatus (Mexico) showed a lower ϕST value (ϕST = 0.302; P = 0.000 ± 0.000) (see Table 2). In an AMOVA comparing the three species and regions 89.21% of the variation was explained by differences between species/regions (d.f 4; P = 0.000) (see Table S2 in Supporting Information).
Haplotype network reconstruction for E. alletteratus resulted in two separate clusters with a complete segregation of the North Atlantic-Mediterranean individuals from the ones from the Tropical East Atlantic (TEA). The north Atlantic cluster showed a star-like pattern with a central haplotype being present in 46 out of 78 individuals, the TEA haplotypes drew a scattered pattern (Fig. 2A). Accordingly, Bayesian Skyline Plots indicates different population dynamics for each location (Fig. 2B, C and D). BSP in NEA haplotypes suggest a recent demographic history with a gradual increase of population size in the last 25,000 years while BSP for NWA individuals indicates a population growth that started around 75,000 years ago. The BSP in the Tropical East Atlantic area reflects a moderate episode of population expansion that started around 125,000 years ago.
Haplotype network of E. alletteratus haplotypes (A). Bayesian Skyline Plots for the North East Atlantic and Mediterranean lineage, NEA (B); North West Atlantic, NWA (C); and the Tropical East Atlantic clade, TEA (D). In the y-axes solid lines represent the median Neτ (τ, generation time) of each population as the product of the effective population size and the generation time for this species, dotted lines indicate de 95% highest probability density limits. X-axes correspond to the time expressed in thousands of years ago (KYA)
As with the mitochondrial marker, CaM showed the lowest gene diversity and nucleotide diversity in the NEA E. alletteratus individuals, while the TEA individuals presented larger genetic diversity (Table 1). Pairwise FST showed significant differentiation between the NEA and the TEA with all alleles private to the respective region. Surprisingly, although significant differences were found between E. affinis and E. lineatus, the FST between them was lower than the expected (Table 3) with several shared alleles. Finally, although the nuclear CaM marker showed a lower value of differentiation than the mtDNA CR, it was still highly significant (FST = 0.654; d.f = 3; P = 0.000).
Phylogenetic analyses and species delimitation
The mtDNA CR phylogenies reconstructed with Bayesian Inference and maximum likelihood clearly segregates the lineages of E. alletteratus into two reciprocally monophyletic clusters that correspond to the entire North Atlantic + Mediterranean and to the Tropical East Atlantic, respectively (Fig. 3). The nodes of these two groups are well supported by bootstrap values and posterior probabilities. These two clusters of E. alletteratus are the sister group to a single well-supported cluster composed by E. affinis + E. lineatus. In this group some of the CR lineages of the two species are lumped together but it can be identified a lineage consisting only of E. lineatus (see Fig. 3 inset). Of the two Bayesian priors, Speciation Yule Process recovered better supported nodes than Coalescence, but in both instances the tree topology for the most internal branches was the same. CaM phylogenetic reconstruction also separates E. alletteratus from the entire North Atlantic + Mediterranean and TEA into two clusters (Fig. 4). For this marker, E. alletteratus from North Atlantic + Mediterranean emerges as the sister taxon of E. affinis + E. lineatus. The E. affinis + E. lineatus alleles are gathered into one cluster with no internal partitioning, with three CaM alleles shared between these two species (but no mtDNA CR haplotype was shared between E. affinis and E. lineatus). This topology is well supported for the two priors tested through Bayesian Inference, although in this case Coalescent constant size recovered well supported nodes (Fig. 4). Unfortunately, maximum likelihood phylogenetic reconstruction for CaM could not recover a congruent topology showing low bootstrap values and several inconsistencies were detected in the allele clustering (data not shown). In accordance with the Bayesian approach, estimation of DA distances for the CR between E. alletteratus from the North Atlantic + Mediterranean vs. TEA (DA = 0.117) was higher than the distance (DA = 0.006) between E. affinis and E. lineatus (Table S3 in Supporting Information).
Bayesian Inference tree under Speciation Yule Process hypothesis for the mtDNA CR marker, nodes support from posterior probabilities (left) and bootstrap values (right) are shown, † indicates three Auxis rochei sequences that were used as outgroup. Green branches correspond to the E. lineatus and E. affinis clade; in the inset black circumferences indicate E. lineatus haplotypes, and grey triangles E. affinis haplotypes. Bars summarize the results for the species delimitation methods tested, with the number of putative species/clusters depicted at the bottom: a Maximum likelihood (M-L) and Bayesian Inference (BI) phylogenetic reconstruction, b Automatic Bar Gap Discovery, c Poisson tree process and multi-rate Poisson tree process, d coalescent reconstruction and Generalized Mixed Yule Coalescent single threshold (cGMYCs), e Yule speciation process and Generalized Mixed Yule Coalescent multiple threshold (yGMYCm), f Yule speciation process and Generalized Mixed Yule Coalescent single threshold(yGMYCs)
Bayesian Inference tree with Coalescent constant size prior for the CaM marker, values on nodes depict support of posterior probabilities. Black circumferences indicate E. lineatus alleles, grey triangles E. affinis alleles, black stars North Atlantic E. alletteratus and white squares South Atlantic E. alletteratus alleles. Shared alleles between E. lineatus and E. affinis are represented with the two symbols (black circumferences and grey triangles). Bars summarize the results of species delimitation methods, with the number of putative species/clusters depicted at the bottom: a Bayesian inference phylogenetic reconstruction, b Automatic Bar Gap Discovery, c multi-rate Poisson tree process, d coalescent reconstruction and Generalized Mixed Yule Coalescent single threshold, e Yule speciation process and Generalized Mixed Yule Coalescent multiple threshold, f Yule speciation process and Generalized Mixed Yule Coalescent single threshold
Results of species delimitation based on mtDNA CR data are summarized in Fig. 3. Excluding the outgroup, the ABGD analysis recovered three clusters (putative species) irrespective of the distance model used. In agreement with the populations analyses presented above, ABGD separated the E. alletteratus individuals from North Atlantic-Mediterranean from those from TEA region as belonging to two different species while grouping E. affinis and E. lineatus into one species. For simple distance some partitions (prior maximal distances [P] ranging from 0.001668 to 0.004642) grouped the dataset into four putative species, with a single haplotype as representative of the fourth species. In addition, PTP and mPTP recovered three species, with one cluster formed by E. affinis and E. lineatus, and two different clusters for individuals from the North Atlantic + Mediterranean region and another for those from the TEA region (P < 0.001, and P < 0.001). Conversely, bPTP retrieved an unrealistic number of species ranging from 39 to 112 and being almost every individual haplotype is identified as a putative species. With GMYC, the most realistic delimitation was obtained under the coalescent prior, in which single threshold performed better than multiple threshold and recovered six clusters as putative species (Confidence Interval [CI] 6–15), with the number of entities increasing to nine if the outgroup is included (CI 9–18). Multiple threshold recovered 14 clusters (CI 13–16) and 18 entities (CI 16–20) (Figure S4 in Supporting Information). On the other hand, GMYC under Yule Speciation Process multiple threshold, while not prove to be better, recovered 13 clusters (CI 13–14) and 18 entities (CI 17–18) (Fig. 1), whereas with single threshold recovered 15 clusters (CI 7–21) and 22 entities (CI 8–28), including the outgroup. Although all the results clearly overestimated the number of species, all tests were statistically significant (P < 0.001).
Species delimitation results obtained with the CaM marker using ABGD tests performed with JC and K80 distances differentiate E. alletteratus from the North Atlantic-Mediterranean locations from the ones from TEA locations, again, one unique cluster was detected for the E. affinis and E. lineatus species (P = 0.001–0.002) (Fig. 4). However, with Jukes Cantor and Kimura models one partition (P = 0.004) clustered all alleles into a single species, and the same results were obtained using simple distance (P = 0.001). PTP reported all alleles except one to belong into a single species (P < 0.001), similarly mPTP reported all alleles as a single species (P < 0.001), while bPTP reported an unrealistic number of species 19 (CI 3–25). In relation to GMYC results, Yule speciation single threshold recovered 2 clusters (CI 1–10) and 2 entities (CI 1–26) while multiple threshold recovered four clusters (CI 1–8) and five entities (CI 1–9), but in both cases without statistical support. On the other hand, GMYC for Coalescent reconstruction single and multiple thresholds recovered 3 clusters (CI 3–9 and CI 3–7, respectively) and 3 entities (CI 3–15 and CI 3–15) being both statistically significant (P < 0.05), GMYC grouped indistinctly individuals of E. affinis and E. lineatus into a single cluster, and split E. alletteratus into two different species, individuals from the North Atlantic-Mediterranean area and individuals from the TEA area.
Discussion
Patterns of genetic differentiation within Euthynnus spp.
In this study, the distribution of genetic variation for E. alletteratus was characterized. To our knowledge this is the largest genetic study on this species in terms of the number of individuals characterized and in geographic coverage. Furthermore, the E. alletteratus variation was compared with the genetic variation of the other two species of the Genus: E. lineatus and E. affinis.
The most salient finding of this paper is the deep genetic divergence within E. alletteratus, which separates the samples from the North Atlantic from the TEA (see below). Previously, Alvarado Bremer & Ely (1999) identified this CR discontinuity when they compared two small samples sizes from NWA and the TEA region. These authors concluded that an amphi-Atlantic pattern of genetic differentiation existed and that the coastal habits of E. alletteratus could account for the absence of trans-Atlantic gene flow that gave rise to the genetic discontinuity. In here this interpretation is partially rejected. The comparison of amphi-Atlantic samples in the North Atlantic reveals that only about 40% of the variation is explained by differences between populations, and that occasional contact between the NWA and the NEA must exist as evidenced by shared haplotypes between the two regions. By contrast, the E. alletteratus from the TEA shows no evidence of gene flow with the NWA and the NEA populations, respectively, (ϕST = 0.866 and 0.918), which is further supported by the reciprocal monophyly of mtDNA and CaM clades. This level of differentiation contrasts dramatically with the shallow index of differentiation (ϕST = 0.302) separating E. affinis from Viet Nam from E. lineatus from Mexico, which is lower than that between the two populations of E. alletteratus at opposite sides of the North Atlantic (ϕST = 0.388). The deep genetic differentiation is corroborated by the different demographic histories suggested by the BSP and network analyses. The NEA population, which include samples from the Mediterranean, seems to have the most recent population history, probably consequence of population expansion after the last glacial period. A similar pattern, population expansion after last glaciation, has been observed in the Mediterranean population of other pelagic species (eg, Atlantic bluefin tuna, Thunnus thynnus; and swordfish, Xiphias gladius Linnaeus, 1758) (Alvarado Bremer et al., 2005). Nevertheless, this assumption should be taken in caution since only one location from the Mediterranean Sea is analysed.
Phylogeny, phylogeographic association, and species delimitations
The phylogeographic association of genetic discontinuities at both the mtDNA and nDNA markers between the North Atlantic and the TEA suggest the presence of two cryptic species of E. alletteratus within the Atlantic Ocean. Both mtDNA CR and CaM data split E. alletteratus individuals into two reciprocally monophyletic clusters supported by bootstrap pseudoreplicates and posterior probabilities (see Figs. 2 and 3). The mtDNA CR divergence between the two groups of E. alletteratus (DA = 11.7%) is nearly 20 times larger than the CR divergence (DA = 0.6%) between E. lineatus (Mexico) and E. affinis (Viet Nam), and similar to the distance that separates E. alletteratus respectively from E. affinis and from E. lineatus (S. Table 3). Further, the level of divergence between the independent lineages of E. alletteratus falls within the range of DA values between species-pairs reported for members of the also cosmopolitan genus Sarda, which range from 11.2 and 17.1% (Viñas et al., 2010). This division of E. alletteratus into two sister taxa was supported by CR data using M-L BI, ABGD and PTP mPTP, and by CAM data using BI, ABGD and cGMYCs&m (see Figs. 2 and 3). The analysis of CR with cGMYCs also identifies E. alletteratus from the TEA as a species separate from the representative samples from North Atlantic, although the latter are also divided into three separate species with no phylogeographic association within that region. By contrast, the results obtained with yGMYCm and yGMYCs using E. alletteratus CR data resulted in the identification of nine and five species, respectively, in the TEA, and two and nine species in the North Atlantic. The analysis of CaM using yGMYCm resulted in the partition of the TEA sample into four species with a fifth grouping the North Atlantic E. alletteratus + E.affinis + E.lineatus as one species. Several authors reported that GMYC tend to over split the individuals in many more clusters than the ones expected (Zhang et al., 2013; da Silva et al., 2018). Long branches and fast evolutionary rates can lead to over split clusters while they are not different species, thus, reflecting population structure rather than different species. The GMYC also present low accuracy in species with large effective population sizes (Esselstyn et al., 2012), consistent with the results obtained here. Thus, it appears that GMYC approaches fail to delimit the processes of speciation and coalescence revealed by CR data, perhaps due to the excessive variability characteristic of this mtDNA segment and attempts to improve the resolving power using mGMYC only increased the number of putative species identified. Nevertheless, this genetic marker has been proved to be marker of choice for delimitating species in the Scombridae family (Viñas et al., 2010; Ollé et al., 2019) with proved higher power of resolution than the traditional COI marker used for DNA barcoding (Viñas & Tudela, 2009). By contrast, the analysis of CaM data using mPTP failed to separate any of the Euthynnus spp. included in this study (Fig. 3). It has been reported before that mPTP and PTP tend to underestimate the number of species (da Silva et al., 2018), which is consistent with the results obtained here. No improvement of species identification is observed for the CaM marker under Yule speciation process ultrametric tree reconstructions. However, for CaM data both single and multiple GMYC approaches under coalescent prior of the phylogenetic reconstructions reported the same three clusters as ABGD and mPTP and PTP mentioned above. As can be seen the different methodologies lead to different conclusions, some of which are highly unrealistic. We advocate that species delimitation methods should include several unlinked markers and different methodologies, including phylogenetic analyses. Then, the most plausible hypothesis is the one shared by most markers and methodologies.
Considering all the results across methods and markers and rationalizing the number of species, we propose that E. alletteratus fishery targets two putative cryptic species that occupy different regions of the Atlantic basin. These results are consistent with the morphological differentiation of E. alletteratus along the western coast of Africa. Gaykov & Bokhanov (2008) compared samples collected from Morocco to Liberia (Central Eastern) against samples collected from Nigeria to Namibia (South-eastern) and found significant differences (P < 0.01) in preanal distance, length of the second dorsal and anal fins base, distance between the caudal fin ends, maximum and minimum body depth, maximum and minimum body width, distance between caudal peduncle keels, and differences in meristic counts in the number of rays in the first dorsal and anal fins. It is very likely that the differences detected in here between the North Atlantic and the TEA samples at the molecular level correspond to the morphological differences identified by Gaykov & Bokhanov, 2008. The North-Tropical Atlantic phylogeographic pattern observed in E. alletteratus fits with the previously defined biogeographic realms (Spalding et al., 2012), that are associated with the North and South Atlantic gyres and the Atlantic Equatorial Current System. This association can be the result of oceanic forces that favoured the reproductive isolation between north and tropical individuals or driven by distinct habitat preferences between species. This association with the oceanic biogeographic realms can help to understand how individuals that are geographically more distant (Tunisia vs Gulf of Mexico) depict lower genetic differentiation than individuals that inhabit closer areas (Portugal vs Senegal). A similar pattern has been observed in other pelagic species such as Delphinus spp. (Amaral et al., 2012) and the swordfish with the use of both mtDNA CR and nuclear markers, including CaM (Alvarado Bremer et al., 1996; Chow et al., 2007). North–South differentiation is supported by the mtDNA CR marker with the split of two clusters of E. alletteratus well supported for bootstrap values and posterior probabilities. Moreover, amphi-Atlantic population differentiation have been detected in other small scombrid species such as Sarda sarda (Bloch, 1793) (Viñas et al., 2010) and Scomber scombrus Linnaeus, 1758 (Rodríguez-Ezpeleta et al., 2016). Additional information is needed to examine the patterns of genetic variation along the eastern coast of Africa, from Morocco to Liberia, which was not sampled in this study. Conversely, in the NWA, the southern portion of its range, from the Caribbean Sea to the northwest coast of South America, needs to be examined.
The high level of differentiation found in E. alletteratus within the Atlantic Ocean contrasts with the shallow genetic divergence that separates E. affinis and E. lineatus collected roughly 15,000 km apart. The DA value (0.6%) and the absence of reciprocal monophyly for either of the two markers is more consistent with population differentiation than that expected for two separate sister species (Table 2 and Table 3; Figs. 2 and 3). Several hypotheses can be invoked to account for this surprising result. First, the range of E. affinis extends from Indian Ocean to Hawaii in the Pacific Ocean, while E. lineatus is primarily found from southern California to Peru in the East Pacific (Fig. 1), although two stray specimens have been collected in the Hawaiian Islands (Collette & Nauen, 1983). Conversely, there are reports of E. affinis captured in areas as far as Baja California and Ecuador, and authors and governmental authorities seems to acknowledge the presence of two different stocks of E. affinis in Western Pacific and Central-Eastern Pacific (Ward, 1995) probably related to a heterogeneous distribution of two highly divergent clades (Jackson et al., 2014). Thus, one possibility is that the sample of E. lineatus from the East Pacific (Mexico) analysed in here included transient E. affinis individuals originating in the Central-Eastern Pacific. A second hypothesis is the small sample sizes characterized in this study compared to those of E. alletteratus. Small sample sizes may result in false negatives, that is the failure to detect differences between two species (Björklund & Bergek, 2009). This explanation, however, is unlikely given the highresolving power of CR data to differentiate among tunas and billfishes (Alvarado Bremer et al., 1997; McKenzie & Alvarado Bremer, 2017). A third hypothesis is incomplete lineage sorting. While reciprocal monophyly is expected between species-pairs, there are several examples of paraphyly due to retention of ancestral polymorphisms or hybridisation in fishes (Campton & Utter, 1985; Smith, 1992; Scribner et al., 2000), and such events have also been reported for marine pelagic species (Chow & Kishino, 1995; Alvarado Bremer et al., 2005; Rooker et al., 2007; Viñas et al., 2010). Given that E. affinis and E. lineatus are one of the youngest lineages upon scombrid species (Santini et al., 2013), the possibility of incomplete lineage sorting needs to be considered. It is worth mentioning that the E. lineatus sample contains a strongly supported cluster of lineages that are private to that sample, which may be support the mixed sample hypothesis, but it is also consistent with the potential of unidirectional gene flow from west to east in the Pacific. Accordingly, a fourth hypothesis needs to be advanced, where E. affinis and E. lineatus are populations of a single species. Traditionally they have been classified as separate species on the basis of differences in meristic counts and morphological characters (Collette & Nauen, 1983). Unfortunately, samples from Mexico were collected under the assumption of belonging to E. lineatus and no meristic counts nor morphological descriptions were recorded. Prior phylogenetic studies using COI data had confirmed the separation of these species, but these studies were based on a single genetic marker and a single individual of E. lineatus (Kumar & Kunal, 2013; Santini et al., 2013). Given that morphological and meristic characters can distinguish E. affinis and E. lineatus individuals, for the moment we should be conservative and keep them separate. However, we recommend a re-evaluation of their status through the genomic characterization of representative samples of both species throughout their range, with a description of each specimen sampled.
Fishery management implications for E. alletteratus
As mentioned in the introduction, no management recommendation for E. alletteratus has been released by ICCAT (ICCAT, 2020). However, for E. alletteratus once we consider the North Atlantic and the TEA regions contain two evolutionary significant units, the regional characterization of these units using the mtDNA CR provides an initial insight into their genetic population structure. Characterization of the North Atlantic region revealed a clear pattern of population substructure, with two genetically distinct populations corresponding to the NWA and the NEA (Table 2). Additionally, based on mitochondrial information from temporal samples, we can acknowledge temporal stability on the genetic differentiation patterns within region. In sum, our results indicate two scales of differentiation in what is now considered a single population with no defined boundaries. The first, supported by the deep evolutionary differences between the North Atlantic and the TEA individuals. The second, based upon the significant differentiation between the Northeast and the Northwest Atlantic regions. Accordingly, for management purposes the populations of E. alletteratus, should be divided into a minimum of two separate units: Tropical East Atlantic and North Atlantic and Mediterranean, with a further consideration of a third unit in the North West Atlantic.
References
Allaya, H., A. Ben Faleh, A. Hattour, M. Trabelsi, & J. Viñas, 2015. Disparate past demographic histories of three small Scombridae (Actinopterygii) species in Tunisian waters. Hydrobiologia 758(1): 19–30.
Alvarado Bremer, J. R. & B. Ely, 1999. Pronounced levels of genetic differentiation between two trans-atlantic samples of little tunny (Euthynnus alletteratus). Collective Volume of Scientific Papers - ICCAT 49: 236–242.
Alvarado Bremer, J. R., J. Mejuto, T. W. Greig, & B. Ely, 1996. Global population structure of the swordfish (Xiphias gladius L.) as revealed by analysis of the mitochondrial DNA control region. Journal of Experimental Marine Biology and Ecology 197: 295–310.
Alvarado Bremer, J. R., I. Naseri & B. Ely, 1997. Orthodox and unorthodox phylogenetic relationships among tunas revealed by the nucleotide sequence analysis of the mitochondrial DNA control region. Journal of Fish Biology 50: 540–554.
Alvarado Bremer, J. R., J. Viñas, J. Mejuto, B. Ely & C. Pla, 2005. Comparative phylogeography of Atlantic bluefin tuna and swordfish: the combined effects of vicariance, secondary contact, introgression, and population expansion on the regional phylogenies of two highly migratory pelagic fishes. Molecular Phylogenetics and Evolution 36: 169–187.
Amaral, A. R., L. B. Beheregaray, K. Bilgmann, D. Boutov, L. Freitas, K. M. Robertson, M. Sequeira, K. A. Stockin, M. M. Coelho & L. M. Möller, 2012. Seascape genetics of a globally distributed, highly mobile marine mammal: the short-beaked common dolphin (genus delphinus). PLoS ONE 7: e31482.
Björklund, M. & S. Bergek, 2009. On the relationship between population differentiation and sampling effort: is more always better?. Oikos 118: 1127–1129.
Campton, D. E. & F. M. Utter, 1985. Natural hybridization between Steelhead Trout (Salmo gairdneri) and Coastal Cutthroat Trout (Salmo clarki clarki) in two puget sound streams. Canadian Journal of Fisheries and Aquatic Sciences 42: 110–119.
Carlsson, J., J. R. McDowell, P. Díaz-Jaimes, J. E. L. Carlsson, S. B. Boles, J. R. Gold & J. E. Graves, 2004. Microsatellite and mitochondrial DNA analyses of Atlantic bluefin tuna (Thunnus thynnus thynnus) population structure in the Mediterranean Sea. Molecular Ecology 13: 3345–3356.
Chow, S. & H. Kishino, 1995. Phylogenetic relationships between tuna species of the genus Thunnus (Scombridae: Teleostei): Inconsistent implications from morphology, nuclear and mitochondrial genomes. Journal of Molecular Evolution 41: 741–748.
Chow, S. & H. Takeyama, 2000. Nuclear and mitochondrial DNA analyses reveal four genetically separated breeding units of the swordfish. Journal of Fish Biology 56: 1087–1098.
Chow, S., S. Clarke, M. Nakadate & M. Okazaki, 2007. Boundary between the north and south Atlantic populations of the swordfish (Xiphias gladius) inferred by a single nucleotide polymorphism at calmodulin gene intron. Marine Biology 152: 87–93.
Collette, B. B., K. E. Carpenter, B. A. Polidoro, M. J. Juan-Jordá, A. Boustany, D. J. Die, C. Elfes, W. Fox, J. Graves, L. R. Harrison, R. McManus, C. V Minte-Vera, R. Nelson, V. Restrepo, J. Schratwieser, C.-L. Sun, A. Amorim, M. Brick Peres, C. Canales, G. Cardenas, S.-K. Chang, W.-C. Chiang, N. de Oliveira Leite, H. Harwell, R. Lessa, F. L. Fredou, H. A. Oxenford, R. Serra, K.-T. Shao, R. Sumaila, S.-P. Wang, R. Watson & E. Yáñez, 2011. High value and long life—double jeopardy for tunas and billfishes. Science 333: 291–292.
Collette, B. B. & C. E. Nauen, 1983. FAO Species Catalogue: Vol. 2 Scombrids of the World. FAO Fisheries Synopsis. http://www.fao.org/docrep/009/ac478e/ac478e00.htm.
da Silva, R., P. L. V. Peloso, M. J. Sturaro, I. Veneza, I. Sampaio, H. Schneider & G. Gomes, 2018. Comparative analyses of species delimitation methods with molecular data in snappers (Perciformes: Lutjaninae). Mitochondrial DNA Part A: DNA Mapping, Sequencing, and Analysis 29: 1108–1114.
Díaz-Arce, N. & N. Rodríguez-Ezpeleta, 2019. Selecting RAD-seq data analysis parameters for population genetics: the more the better? Frontiers in Genetics 10: 1–10.
Drummond, A. J., M. A. Suchard, D. Xie & A. Rambaut, 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution 29: 1969–1973.
Ely, B., J. Viñas, J. R. Alvarado Bremer, D. Black, L. Lucas, K. Covello, A. V. Labrie & E. Thelen, 2005. Consequences of the historical demography on the global population structure of two highly migratory cosmopolitan marine fishes: the yellowfin tuna (Thunnus albacares) and the skipjack tuna (Katsuwonus pelamis). BMC Evolutionary Biology 5: 1–9.
Esselstyn, J. A., B. J. Evans, J. L. Sedlock, F. A. A. Khan & L. R. Heaney, 2012. Single-locus species delimitation: a test of the mixed yule-coalescent model, with an empirical application to Philippine round-leaf bats. Proceedings of the Royal Society B: Biological Sciences 279: 3678–3686.
Excoffier, L. & H. E. L. Lischer, 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10: 564–567.
Excoffier, L., P. E. Smouse & J. M. Quattro, 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491.
Gaykov, V. Z. & D. V. Bokhanov, 2008. The biological characteristic of Atlantic black skipjack (Euthynnus alletteratus) of the eastern Atlantic ocean. Collective Volume of Scientific Papers - ICCAT 62: 1610–1628.
Gernhard, T., 2008. The conditioned reconstructed process. Journal of Theoretical Biology 253: 769–778.
Huelsenbeck, J. P. & J. J. Bull, 1996. A likelihood ratio test to detect conflicting phylogenetic signal. Systematic Biology 45: 92–98.
ICCAT, 2020. Compendium management recommendations and resolutions adopted by ICCAT for the conservation of Atlantic tunas and tuna-like species: 1–447.
Jackson, A. M., Ambariyanto, M. V. Erdmann, A. H. A. Toha, L. A. Stevens & P. H. Barber, 2014. Phylogeography of commercial tuna and mackerel in the Indonesian Archipelago. Bulletin of Marine Science 90: 471–492.
Juan-Jordá, M. J., I. Mosqueira, A. B. Cooper, J. Freire & N. K. Dulvy, 2011. Global population trajectories of tunas and their relatives. Proceedings of the National Academy of Sciences of the United States of America National Academy of Sciences 108: 20650–20655.
Juan-Jordá, M. J., I. Mosqueira, J. Freire & N. K. Dulvy, 2013. The conservation and management of tunas and their relatives: setting life history research priorities. PLoS ONE 8: e70405.
Kapli, P., S. Lutteropp, J. Zhang, K. Kobert, P. Pavlidis, A. Stamatakis & T. Flouri, 2017. Phylogenetics Multi-rate Poisson tree processes for single-locus species delimitation under maximum likelihood and Markov chain Monte Carlo. Bioinformatics 33: 1630–1638.
Kearse, M., R. Moir, A. Wilson, S. Stones-Havas, M. Cheung, S. Sturrock, S. Buxton, A. Cooper, S. Markowitz, C. Duran, T. Thierer, B. Ashton, P. Meintjes & A. Drummond, 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 19: 34–17.
Kekkonen, M. & P. D. N. Hebert, 2014. DNA barcode-based delineation of putative species: efficient start for taxonomic workflows. Molecular Ecology Resources 14: 706–715.
Kumar, G. & S. P. Kunal, 2013. Historic demography and phylogenetic relationship of Euthynnus species based on COI sequence analyses. International Journal of Bioinformatics Research and Applications 9: 547–555.
Kumar, G., S. P. Kunal, M. R. Menezes & R. M. Meena, 2012. Single genetic stock of kawakawa Euthynnus affinis (Cantor, 1849) along the Indian coast inferred from sequence analyses of mitochondrial DNA D-loop region. Conservation Genetics 1119–1131.
Kumar, S., G. Stecher, K. Tamura & J. Dudley, 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets downloaded from. Molecular Biology and Evolution 33: 1870–1874.
Laconcha, U., M. Iriondo, H. Arrizabalaga, C. Manzano, P. Markaide, I. Montes, I. Zarraonaindia, I. Velado, E. Bilbao, N. Goñi, J. Santiago, A. Domingo, S. Karakulak, I. Oray & A. Estonba, 2015. New nuclear SNP markers unravel the genetic structure and effective population size of albacore tuna (Thunnus alalunga). PLoS ONE 10: 1–19.
Leigh, J. W. & D. Bryant, 2015. POPART: Full-feature software for haplotype network construction. Methods in Ecology and Evolution 6: 1110–1116.
Lucena-Frédou, F., B. Mourato, T. Frédou, P. G. Lino, R. Muñoz-Lechuga, C. Palma, A. Soares & M. Pons, 2021. Review of the life history, fisheries, and stock assessment for small tunas in the Atlantic Ocean. Reviews in Fish Biology and Fisheries 31: 709–736.
McKenzie, J. L. & J. R. Alvarado Bremer, 2017. Genetic identification of istiophorid larvae from the Gulf of Mexico based on the analysis of mitochondrial DNA control region sequences. Journal of Fish Biology 90: 1070–1079.
Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
Nei, M. & F. Tajima, 1981. DNA polymorphism detectable by restriction endonucleases. Genetics Genetics 97: 145–163.
Nei, M., F. Tajima & Y. Tateno, 1983. Accuracy of estimated phylogenetic trees from molecular data. Journal of Molecular Evolution 19: 153–170.
Ollé, J., D. Macías, S. Saber, M. J. Gómez-Vives, N. Pérez-Bielsa & J. Viñas, 2019. Genetic analysis reveals the presence of frigate tuna (Auxis thazard) in the bullet tuna (Auxis rochei) fishery of the Iberian Peninsula and the western-central Mediterranean Sea. Bulletin of Marine Science. https://doi.org/10.5343/bms.2018.0049.
Palumbi, S. R., 1994. Genetic divergence, reprodutive isolation and marine speciation. Annual Review of Ecology, Evolution, and Systematics 25: 547–572.
Pascual, M., B. Rives, C. Schunter & E. Macpherson, 2017. Impact of life history traits on gene flow: a multispecies systematic review across oceanographic barriers in the Mediterranean Sea. PLoS ONE 12: 1–20.
Pons, J., T. G. Barraclough, J. Gomez-Zurita, A. Cardoso, D. P. Duran, S. Hazell, S. Kamoun, W. D. Sumlin & A. P. Vogler, 2006. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Systematic Biology 55: 595–609.
Qiu, F., A. Kitchen, P. Beerli & M. M. Miyamoto, 2013. A possible explanation for the population size discrepancy in tuna (genus Thunnus) estimated from mitochondrial DNA and microsatellite data. Molecular Phylogenetics and Evolution 66: 463–468.
Rambaud, A., 2012. FigTree. University of Edinburgh. http://tree.bio.ed.ac.uk/software/figtree/.
Rambaut, A., A. J. Drummond, D. Xie, G. Baele & M. A. Suchard, 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Systematic Biology 67: 901–904.
Rodríguez-Ezpeleta, N., I. R. Bradbury, I. Naki Mendibil & P. Alvarez, 2016. Population structure of Atlantic mackerel inferred from RAD-seq-derived SNP markers: effects of sequence clustering parameters and hierarchical SNP selection. Molecular Ecology Resources 16: 991–1001.
Rooker, J. R., J. R. Alvarado Bremer, B. A. Block, H. Dewar, G. De Metrio, A. Corriero, R. T. Kraus, E. D. Prince, E. Rodriguez-Marin & D. H. Secor, 2007. Life history and stock structure of atlantic Bluefin Tuna (Thunnus thynnus). Reviews in Fisheries Science 15(4): 265–310.
Rozas, J., A. Ferrer-Mata, J. C. Sánchez-DelBarrio, S. Guirao-Rico, P. Librado, S. E. Ramos-Onsins & A. Sánchez-Gracia, 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Molecular Biology and Evolution 34: 3299–3302.
Saber, S., J. Ortiz de Urbina, P. G. Lino, M. Gómez-Vives, R. Coelho, R. Muñoz-Lechuga & D. Macías, 2018. Biological aspects of little tunny Euthynnus Alletteratus from Spanish and Portuguese waters. Collective Volume of Scientific Papers, ICCAT 75: 95–110.
Santini, F., G. Carnevale & L. Sorenson, 2013. First molecular scombrid timetree (Percomorpha: Scombridae) shows recent radiation of tunas following invasion of pelagic habitat. Italian Journal of Zoology Taylor & Francis 80: 210–221.
Scribner, K. T., K. S. Page & M. L. Bartron, 2000. Hybridization in freshwater fishes: a review of case studies and cytonuclear methods of biological inference. Reviews in Fish Biology and Fisheries 10: 293–323.
Smith, B. L., C. P. Lu, B. García-Cortés, J. Viñas, S. Y. Yeh & J. R. Alvarado Bremer, 2015. Multilocus Bayesian estimates of intra-oceanic genetic differentiation, connectivity, and admixture in Atlantic Swordfish (Xiphias gladius L.). PLoS ONE. https://doi.org/10.1371/journal.pone.0127979.
Smith, G. R., 1992. Introgression in fishes: significance for paleontology, cladistics, and evolutionary rates. Systematic Biology 41: 41–57.
Spalding, M. D., V. N. Agostini, J. Rice & S. M. Grant, 2012. Pelagic provinces of the world: a biogeographic classification of the world’s surface pelagic waters. Ocean and Coastal Management 60: 19–30.
Stephens, M., N. J. Smith & P. Donnelly, 2001. A new statistical method for haplotype reconstruction from population data. The American Journal of Human Genetics Cell Press 68: 978–989.
Theisen, T. C., B. W. Bowen, W. Lanier & J. D. Baldwin, 2008. High connectivity on a global scale in the pelagic wahoo, Acanthocybium solandri (tuna family Scombridae). Molecular Ecology 17: 4233–4247.
Thompson, J. D., D. G. Higgins & T. J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680.
Viñas, J., J. R. Alvarado Bremer & C. Pla, 2010. Phylogeography and phylogeny of the epineritic cosmopolitan bonitos of the genus Sarda (Cuvier): inferred patterns of intra- and inter-oceanic connectivity derived from nuclear and mitochondrial DNA data. Journal of Biogeography 37: 557–570.
Viñas, J. & S. Tudela, 2009. A validated methodology for genetic identification of tuna species (genus Thunnus). PLoS ONE 4: e7606.
Waples, R. S., 1998. Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. The American Genetic Association 89: 438–450.
Waples, R. S., P. M. Grewe, M. W. Bravington, R. Hillary & P. Feutry, 2018. Robust estimates of a high Ne/N ratio in a top marine predator, southern bluefin tuna. Science Advances 4: eaar7759.
Ward, R. D., 1995. Population genetics of tunas. Journal of Fish Biology 47: 259–280.
Wright, S., 1965. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution [Society for the Study of Evolution, Wiley] 19: 395–420.
Yule, G. U., 1924. A mathematical theory of evolution, based on the conclusions of Dr. JC Willis, FRS. Philosophical Transactions ofthe Royal Society ofLondon Series B, Containing Papers of a Biological Character 213: 21–87.
Zhang, J., P. Kapli, P. Pavlidis & A. Stamatakis, 2013. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29: 2869–2876.
Acknowledgements
“This work was carried out under the provision of the ICCAT Small Tunas Year Program (SMTYP). The contents of this paper do not necessarily reflect the point of view of ICCAT, which has no responsibility over them, and in no ways anticipate the Commission’s future policy in this area.” We would like to thank all the members of the ICCAT Small Tuna Species Group for their comments during the meetings that really helped in the construction of this study.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature.
Author information
Authors and Affiliations
Contributions
JO and LVV performed the wet lab work. JO analysed the data and drafted the manuscript. JAB, NS, RMA and JV supervised the genetic analysis and contributed to the manuscript. GC, TYD, GH, PGL, RML, FNS and NCD provided the samples and contributed to the manuscript. JO and JV conceived the study.
Corresponding author
Additional information
Handling editor: Cécile Fauvelot
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ollé, J., Vilà-Valls, L., Alvarado-Bremer, J. et al. Population genetics meets phylogenetics: new insights into the relationships among members of the genus Euthynnus (family Scombridae). Hydrobiologia 849, 47–62 (2022). https://doi.org/10.1007/s10750-021-04707-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-021-04707-6