Abstract
The new method of biological assessment in flowing waters—NoMBSI (Non-lethal Method for Benthos Sampling and Identification) is presented. To determine the amount of information (informativity), which is possible to obtain during a sample analysis, the results of three procedures were compared (NM-the new one based on 3D digital image analysis, NC-control procedure based on microscopic identification and G, which is the traditional method based on samples collected with Günther sampler). Procedures were applied for the samples collected at 27 sites in North-eastern and Eastern Poland. The NM was found to provide for the correct recognition of an average of 45% of taxa present in samples. Taxa of small size and lower abundance were frequently failed to be identified. However, no significant differences between NM and NC in terms of raw data on taxonomic composition were found, while the results of G procedure were significantly different when compared with both other procedures. Values of commonly used biotic and diversity indices calculated on the basis of those procedures were compared using correlation. The level of identification achieved sufficed for the proper determination of water quality classes at all of the sites sampled.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Macroinvertebrates have many features which make them nearly ideally suitable for biomonitoring (Bonada et al., 2006). This has ensured the popularity of quality assessment methods featuring benthic environments and application of some of these methods in national routine monitoring programs (e.g., De Pauw & Vanhooren, 1983; Barbour et al., 1999; Davies, 2000; Birk et al., 2010). However, most of these assessment procedures using benthic animals entail the same consecutive stages of (a) sampling, (b) sample preservation, (c) sample processing—taxa identification and counting—and (d) data interpretation (e.g., Barbour et al., 1999; AQEM Consortium, 2002). This approach inevitably leads to the death of all animals in a sample. Furthermore, in a sense, this stands in opposition to several national and international agreements, polices and other statements, or documents, which prohibit actions that put protected, rare and threatened animal species, or biological diversity in general, under pressure. The scale of the problem is made clear when it is noted that a single sampling episode that is in line with the Polish modification to the BMWP method (Fleituch et al., 2002; Kownacki & Soszka, 2004) or STAR/AQEM (AQEM Consortium, 2002) typically involves the deaths of more than 2,500, or even more than 5,000 individual animals, respectively. Moreover, sub-sample of a representative number of individuals is permitted where a sample proves to be very abundant. Where considerations are restricted to such a sub-sample, no use is found for the remaining individuals in subsequent phases of assessment (e.g. Kornijow & Lechowska 2002; Kownacki et al., 2002; Verdonschot, 2006). In this way, traditional methods of biological monitoring not only fail to protect fragments of valuable natural resources adequately, but also become questionable on ethical grounds. Unfortunately, however, the long history of the elaboration of assessment methods (Cairns & Pratt, 1993; Bailey et al., 2004) has not yet seen the emergence of non-lethal procedures using macroinvertebrates.
It is clear that the sample preservation and processing stages of assessment procedures are crucial in terms of lethality for invertebrates. Meanwhile, the mortality of sampled animals and the assessment efficiency of the different biotic indices are independent of each other. This idea underlies development of a new method known as NoMBSI (Non-lethal Method for Benthos Sampling and Identification) described in this article. In general, NoMBSI includes the same stages that have been employed traditionally (i.e., sampling, sample preservation, and taxa identification). However, the particular stages differ in their details. Sample preservation and taxa identification have been made subject to far-reaching modifications, in that “digital preservation” and image analysis take the place of sample preservation in alcohol and taxa identification using a stereoscopic microscope. Since the primary focus of this procedure is on non-lethal taxa identification, there is no new means of data interpretation (e.g., no original biotic index) detailed here, but rather testing on the basis of already existing indices. This article contains detailed description of the non-lethal procedure, as well as the preliminary results obtained from its testing.
The main aim of the work described here was thus to embrace the increasing need for a reduced pressure on assemblages of benthic macroinvertebrates and to test the assumption that information sufficient for the correct assessment of the ecological status of flowing waters may be obtained without resort to the mass killing of benthic animals.
Materials and methods
Study area
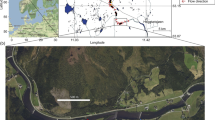
Field sampling was carried out in 2012 and 2013 in north-eastern and eastern Poland (Fig. 1). The study area represents lowland and highland landscape and is classified into seven physico-geographical macroregions of the Mazurian Lakeland, the North Mazowsze Lowland, the Central Mazowsze Lowland, the South Podlasie Lowland, the Western Polesie and the Lublin Upland (Kondracki, 2002). In line with WFD requirements, sampled waters were classified into nine different abiotic types and five biocoenotic types (Bis & Mikulec, 2013). Since study sites are also used as sampling points by Voivodship Inspectorates for Environmental Protection (WIOŚ), information on ecological status was obtained from the online reports published annually by these institutions. In total, 27 sampling sites differing in terms of their abiotic type and ecological status were selected for study (Table 1).
The collection, preservation, and identification of benthic macroinvertebrates
Achievement of the study’s objective entailed the comparison of datasets obtained using different methods of sampling, preservation, and identification. Specifically, two procedures appropriate in biological assessment were applied.
The first procedure (in accordance with Polish official modification of BMWP method) entailed multi-habitat sampling of a quantitative nature—using a Günther bottom sampler, or else a semi-quantitative nature—using hand netting for non-bottom habitats, as advocated by Fleituch et al. (2002), and Kownacki & Soszka (2004). A sample consisted of three standard Günther sampler units and five standard hand-net sweeps (total sampling area ca. 0.367 m2). A single sample for each site was taken in June (only sample at site 21 was not taken using this procedure due to bridge reconstruction and thus bottom disturbance), with collected material being drained off through a 1 mm mesh net and then preserved in 75% alcohol. Animals were then sorted under a stereoscopic microscope in the laboratory. The whole procedure described in this paragraph is indicated in the text below using the letter G.
The second procedure suggested in the case of NoMBSI is based on artificial substrate samplers (AS) placed in a riffle (e.g., De Pauw et al., 1986; Czerniawska-Kusza, 2004). The single AS was constructed out of a set of ten combined, polypropylene cubic test tube racks with a total area of ca. 1 m2. The whole set was placed into a potato bag with a mesh size of ca. 10 mm, and weighted down with stones (Online Resources, Fig. S1). Single AS setups were put in place in June at the same day as G sampling (only at sites 16 and 17 two sets were used), and then left for a period of 11–14 weeks until late summer. Following removal of the AS, animals present were flushed out, sieved (in 1 mm mesh net), placed in a photographic tray (Online Resources, Fig. S2) of the transportable photographic studio (3DS), and supplied with a water to the level of ca. 2 cm (overall volume of digitalize sample was ca. 2.5 L). With the aid of 3DS, collected samples were digitalized in the form of 3D images with a resolution of 3,648 × 2,736 pixels. The 3DS consists of a non-transparent plastic box (width × height × length: 600 × 900 × 400 mm) equipped with a photographic tray on the bottom and a set of two digital photographic cameras attached to the upper cover. In order to synchronize cameras, shutters of both are triggered simultaneously by infrared remote control. Flash lamps serve as a source of light (Online Resources, Fig. S3). The total time spent preparing one sample in the field (up to the time photographs are taken) was in the range of 30–60 min. Most of the animals were still alive when we finished digitalization. After digitalization, collected material was preserved in 75% alcohol and sorted in the laboratory under a stereoscopic microscope, serving as control for non-lethal preservation and identification. Taxa identification was carried out by analysing images with StereoPhoto Maker version 4.36 (http://stereo.jpn.org/eng/stphmkr/index.html). Digital images of samples were analysed independently by two identifiers (A and B), in order for the impact of the person engaged in identification to be estimated. The results provided by NoMBSI are indicated using the letters NM in the text below, while the control procedure is identified by the letters NC.
Since family-level seems sufficient for reliable ecological status assessments using most procedures (e.g., Armitage et al., 1983; Hilsenhoff, 1988; Smith et al., 1999), benthic macroinvertebrates sampled were only identified to this level. Exceptions were Oligochaeta, Tricladida, and Hydrachnellae, which were identified to a level higher than family. For the purposes of the study, identification was achieved in line with the key for macroinvertebrates developed by Kołodziejczyk & Koperski (2000).
Description of NoMBSI data in comparison to NC procedure
To compare raw data (pres/abs and taxa abundance) derived from the non-lethal and control procedures, measurements were made in respect of parameters presented below:
-
The percentage of properly identified taxa per sample,
-
The percentage of incorrectly identified taxa per samples (taxa identified in the NM samples but not found in particular NC samples were treated as identified incorrectly),
-
The frequency of occurrence of unidentified taxa,
-
The frequency of occurrence of incorrectly identified taxa,
-
A list of percentage of samples with unidentified taxa,
-
Average numbers of unidentified taxa in control samples,
-
Ratio of ease of taxon identification (recognizability), calculated for all taxa in given samples, as:
$$\left[ {\left( {{{N}}_{{d}} /{\text{N}}_{{p}} } \right)_{ 1} + \left( {{{N}}_{{d}} /{\text{N}}_{{p}} } \right)_{ 2} + \cdots + \left( {{{N}}_{{d}} /{{N}}_{{p}} } \right)_{{n}} } \right]/{{n,}}$$where N d is the number of properly detected individuals, N p the number of individuals present in a sample, and n the number of samples.
Values for recognizability ratio supply information on the frequency and number of correctly identified taxa and individuals. However, in the case of very low abundant taxa, values for recognizability seem to be underestimated. Obviously the amount of sediment might influence the effectiveness of taxa identification and hence the proportion of the photographic tray covered by sediments was thus measured for all samples by ImageJ computer software (Schneider et al., 2012). Potential differences in datasets prepared by the two identifiers were also taken into consideration.
Since the percentages for correctly and incorrectly identified taxa differed greatly between samples, it was decided to test the strength of particular parameters as potentially influencing factors. Six of these were selected, and classified as categorical factor (identifier effect) or continuous (gradient) parameters (number of taxa per sample, number of individuals per sample, ease of taxon identification per sample, amount of sediment per sample, order in which sample images were analysed). The significance of the influence exerted by categorical factor was tested using Mann–Whitney U and Wilcoxon signed-rank tests (the effect of the identifier where paired and unpaired data were used). The significance of the impact of factors from the second group (continuous parameters) was in turn tested using Multiple Regression analysis.
Informativity of NoMBSI in comparison to NC and G procedures
It is possible to obtain information on resemblances between sampling sites (e.g., polluted and unpolluted) using a similarity ratio determined on the basis of presence/absence data and taxa abundances. To compare differences between similarity matrices created using different methods, nonparametric analysis of variance (PERMANOVA) with Bray-Curtis distance as a similarity measure was used. Comparisons were made for datasets obtained using all procedures (G, NM, and NC). Data sets for NM obtained by the two different identifiers were treated independently. Because of differences in sampling and taxa identification methods, raw datasets were standardized as proportional values per sample. All datasets were also arcsine–transformed prior to analysis.
Discriminant function analysis (DFA) was performed to test the efficiency of ecological quality assessment, on the basis of information obtained from datasets involving NoMBSI and the two other procedures (G and NC). Due to the small and uneven numbers of sites classified to particular levels of ecological status, the 5-point scale of the WFD classification scheme was transformed into a 3-point scale by pooling quality classes together (“high” and “good” into “good”, and “poor” and “bad” into “bad”). Data obtained by the two identifiers were treated as independent measurements of NM and gathered into a single dataset. Prior to analysis, datasets were standardized as percentage values per sample.
Five widely used or nationally applied biotic indices plus the Shannon diversity index were chosen, and calculated for all procedures:
-
Polish Multimetric Index—MMI PL (Bis & Mikulec, 2013),
-
Biological Monitoring Working Party in Polish modification—BMWP-PL (Armitage et al., 1983; Kownacki & Soszka, 2004),
-
Average Score Per Taxa—ASPT (Armitage et al., 1983).
-
Percentage of Ephemeroptera, Plecoptera, and Trichoptera families—[%] EPT (Plafkin et al., 1989),
-
Family Biotic Index—FBI (Hilsenhoff, 1988),
-
Shannon diversity index—H (Shannon, 1948).
Values of all indices from the list above were than correlated between NM, G, and NC. Data obtained by the two identifiers (A and B) were treated as independent measurements of NM gathered into one dataset.
The analysis were carried out using STATISTICA software package (v. 10.0; StatSoft, Inc., 2011), and PAST v. 2.17 for PERMANOVA.
Results
Several substrates, i.e., eight artificial substrates, were lost or damaged while exposed at sampling sites. In total, 26 samples collected with G procedures and 21 samples collected NM procedures were obtained. Twenty-one samples of NC were collected simultaneously.
Description of NoMBSI data in comparison to NC procedure
In total, 71 taxa were present in the NC samples, while 50 and 53 taxa were identified by identifier A and B, respectively, when the NM procedure was used (63 taxa when lists were combined). There were considerable differences between taxa in regard to the percentages of samples in which they were or were not identified (Fig. 2). Almost 96% of all taxa went unrecognized at least once when the NM procedure was applied. Furthermore, 22 of these taxa were not recognized even once, while for the other 46 the frequencies of recognition varied between 10 and 93%. In general, these unrecognized taxa were usually characterized by low or very low abundance (meaning no more than ten individuals in more than 77% of cases—Fig. 3a).
The percentages of properly identified taxa differed between samples. On average, 42 and 48% of the taxa present in a sample were identified correctly by identifiers A and B, respectively (Fig. 4a). Forty and 41 taxa were identified properly at least once by identifiers A and B, respectively (49 for the two considered together).
The percentages of incorrectly identified taxa varied between samples. On average, they were 23 and 20% for identifiers A and B, respectively (Fig. 4b). On the other hand, abundances of particular incorrectly identified taxa were low, with only a single individual being found in more than 58% of cases (Fig. 3b). Forty-six out of 63 taxa were identified incorrectly at least once using NoMBSI.
The values for the ratio of recognizability varied between taxa (Fig. 5). For 49 of them, values for the ratio were higher than 0, unlike in the case of the remaining 22. For six taxa the value of the ratio was 0.5 or more.
The impact of an identifier had a significant influence only on the percentages of correctly identified taxa (Table 2), but just if samples were treated as joint (paired) ones, then one of the identifiers properly identified more taxa than the second one, depending on the particular sample. The ratio of ease of taxon recognition and number of taxa in particular samples were factors that influenced correctness of identification significantly in multiple regression models. The model explained 59% of total variance. There were no significant factors from the two groups in terms of incorrectness of identification.
Informativity of NoMBSI in comparison to NC and G procedures
The analysis showed statistically significant differences between similarity matrices when all three procedures were compared (F = 2.325, P = 0.0005). The similarity matrix for sampling sites obtained using the G procedure differed significantly from those obtained using the other two procedures. However, when NoMBSI and the control were compared, no significant differences between similarity matrices were noted. The same result was found for data obtained by the two different identifiers, also when the two were compared with each other.
It was possible for all samples to be classified properly to one of the three quality classes, regardless of which procedure was used to gather information on the taxonomic composition of the macroinvertebrate assemblages (Fig. 6a–c). It must be emphasized that different variables were important in explaining total variability in datasets obtained using the different procedures. For procedure G, the percentages for Ancylidae and Gammaridae positively and most strongly correlated with DFA axis 1 (83% of explained variability) while Bithynidae and Asellidae positively correlated with DFA axis 2 (17% of explained variability) (Fig. 6a). In the case of NC, 100% of total variability was explained by axis 1 (Fig. 6b). Percentages for Ancylidae and Athericidae were negatively associated with both axes. In the case of NoMBSI, axis 1 explained 100% of the variability and were correlated positively and most strongly with Gyrinidae and Dytiscidae (Fig. 6c).
Results of discriminant function analysis (DFA) for different procedures of sampling and identification a traditional procedure (G), b control procedure for NoMBSI (NC), c NoMBSI procedure (NM). Taxa which percentages most strongly correlated with both discriminant functions are also presented on the plots. Samples were categorized as one of three quality classes: gray circles good, white circles moderate, black circles poor/bad. It was possible to accurately discriminate quality classes of all study sites with all procedures
Correlations between indices values varied depending on compared methods (Table 3).When G and NC methods were compared, only two indices ([%] EPT and FBI) showed significant but very weak correlations between both methods ([%] EPT; R 2 = 0.166, P = 0.009 and FBI; R 2 = 0.202, P = 0.004). Similar results were observed when G and NM methods were compared, with only ASPT and FBI indices proving significant but very weak correlations between both procedures (R 2 = 0.125, P = 0.024 and R 2 = 0.207, P = 0.003, respectively). All indices were significantly correlated between methods when NM and NC methods were compared, though the strengths of the correlations differed between indices. The weakest correlation was observed for BMWP (R 2 = 0.235, P = 0.002), the strongest for [%] EPT (R 2 = 0.657, P = 0.000). The R2 values in remaining cases ranged between 0.545 and 0.557.
Discussion
The main feature of NoMBSI is that the procedure is selective, i.e., focused only on some taxa. First, the artificial substrate (AS) setups being placed in riffles results in colonization by rheophilous taxa. Despite quantitative and qualitative differences between different microhabitats in a river, and their effects on the composition of macrobenthic assemblages (e.g., Álvarez-Cabria et al., 2011), the taxa collected using AS provide sufficient information for degradation to be assessed (De Pauw et al., 1986). On the other hand, less selective methods of sampling (like the multi-habitat sampling procedure—G method) may increase taxa diversity in samples, including easy-to-identify indicative taxa, what might be really useful for NoMBSI. Unfortunately, most of the samples collected with the G method contained big amount of organic and inorganic matter and thus identification of animals through image analysis failed. Another drawback is the high risk of losing of AS due to rapid increase in water current speed or human activity. The former can be especially significant in case of fast flowing streams (e.g., mountain ones) and much of attention should be paid while constructing AS and fixing it at sampling site.
The use of “digital preservation” is associated with increased selectivity. Nevertheless, nearly 70% of the taxa present across samples were identified correctly at least once, and identification of only those animals that are readily recognized does prove sufficient for correct assessment of ecological status. It must also be strongly emphasized that the task this procedure involves is confined to the mere detection of taxa, while abundance is only of secondary importance. Moreover, the use of biodiversity measures based on all macroinvertebrates is often criticized (e.g., Jones, 2008; Koperski, 2011). For instance, Odum et al. (1979) noted that the relationship between the number of macroinvertebrate specimens and organic pollution could be curvilinear rather than linear. While values of indices tested in this article were rather weakly correlated when comparing lethal and non-lethal methods, we conclude that a new biotic index must be created in order to improve efficiency in ecological status discrimination. The biotic index should be designed with account taken of known differences between lists of taxa obtained in accordance with NoMBSI and the lists employed in traditional procedures. The selection of taxa included in the index could be based inter alia on the ratio of ease of taxa recognition. Furthermore, many of the biotic indices currently in use take into account only part (if in most cases a dominant part) of the taxa present in a sample. Good examples here are the EPT (Plafkin et al., 1989), BMWP (Armitage et al., 1983) APODEMAC (Koperski & Golub, 2006) and FBI (Hilsenhoff, 1988) indices.
The number of particular taxa present undoubtedly affects their detectability. Most taxa with a low frequency of identification were also characterized by low abundance, unlike the most frequently recognized taxa. However, this relationship is not linear for all taxa, and at this point the body size of specimens should be also considered. In case of NoMBSI this would seem to be of particular importance, as taxa attaining higher maximum body sizes were detected more frequently. What is more, specimens of some taxa were not identified with the required accuracy due to small body size. Such a correlation is also to be observed for samples collected and identified in the traditional way. For example, smaller larvae of the EPTO (Ephemeroptera, Plecoptera, Trichoptera, and Odonata) group can only be identified at higher taxonomic level (Orlofske & Baird, 2013). Another urgent problem is the potential confusion of some taxa with similar ones (e.g., Libellulidae, Limonidae, Tipulidae, and Polycentropodidae). This issue might be of big importance, in particular for taxa valuable in terms of degradation assessment. In this regards, removal of the most confusing taxa from indicative inference should be considered.
The correctness and incorrectness of taxa identification are both crucial for NoMBSI, and influence the effectiveness of the degradation assessment markedly. It was found the identities of the identifiers only influenced correctness of taxa recognition significantly when samples were treated as related. This may reflect different researchers being more specialized in the identification of different groups of organisms. As a result, the presence of certain taxa in samples has an impact on the percentage of those identified correctly. On the other hand, this may also depends on the individual abilities of the identifier when it comes to the recognition of shapes and colors, or differences in spatial imaging. Moreover, the incorrectness of taxa recognition seems a largely random process susceptible to the influence of individual experience with this type of image analysis. Then again, some of the incorrectly identified animals could be present in NC samples, only to be damaged and/or go missing during sorting in the laboratory with a view to preservation and storage (e.g., Keizer-Vlek et al., 2011). Still, some differences in taxonomic composition between identifiers can be neglected for degradation assessment purposes due to the fact that the particular quality classes are usually described as a range of values and not a single value (e.g., Bis & Mikulec, 2013). Koperski (2014) showed that with the use of NoMBSI correct discrimination of ecological status was possible despite some differences in taxa recognition between identifiers of similar experience.
Raw data on community structure is usually obtained in the form of a matrix (sites as rows × number of individual taxa as columns), while in the modern biological monitoring of freshwater habitats discriminated sites are compared with the least-degraded (reference) sites (e.g., Norris, 1995; Directive 2000/60/EC). The mathematical procedure for comparing samples is often based on measures of similarity (Norris, 1995). The similarity data matrix of NoMBSI was not significantly different from that of the control procedure, this showing clearly NoMBSI allows for the obtainment of information about similarities between sites comparable to procedures requiring killing of invertebrates. As anticipated, significant differences between the NoMBSI and G procedures as well as between NC and G were found. Firstly, differences in taxonomic composition are partly determined by phenology (e.g., De Pauw et al., 1986). Moreover, artificial substrates are more selective than the procedure for multi-habitat sampling (Lenat, 1988). Similarly, values for tested biotic indices were least correlated between G and NC, as well as between G and NM, while the values for all indices were found to correlate when NM and NC were compared.
Despite the differences between the tested procedures in terms of the occurrence and abundance of individual taxa, the differentiation of all sampling sites was found to be correct. This result supports our assumption that only taxa with high indicative value, and not all taxa present in a sample, should be taken into consideration. Taxa most strongly associated with the explanatory axes differed between procedures, but all are nonetheless listed as indicator taxa of the BMWP-PL index (Kownacki & Soszka, 2004).
In modern ecology, the tendency to include DNA-based approach of taxa identification for biological assessment can be seen (Pfrender et al. 2010). However, while molecular genetics might be really helpful in species identification (e.g., Sweeney, 2011), it fails to solve the problem of necessity of killing of the animal. The procedure proposed here is an alternative. To the best of our knowledge, the NoMBSI method is the first of its kind. We do agree it still needs some improvements to increase recognizability of taxa, as in some of the samples it is low. Although Rapid Field Assessment Methods might seem somewhat similar (e.g., Hilsenhoff 1988), this approach is not taken very commonly, as its application is confined to highly experienced specialists, and any further verification is rendered impossible. In contrast, NoMBSI has a high potential for use in rapid assessment methods, where sorting of animals in the course of fieldwork and further selection of their specified number (subsample) should increase recognizability of taxa and thus other related parameters. The above modifications thereby can be used in case of any kind of sampling procedures. As the quality of cameras is undoubtedly crucial for NoMBSI, the use of much higher resolution cameras should help in identification of taxa which are small and easy to confuse. Furthermore, restricted area of research and the use of only 9 out of 26 abiotic types present in Poland can obscure the results. Thus, more study sites spanning both larger area and more habitat types will need to be considered.
NoMBSI can be also suggested as an alternative for traditional methods in terms of time- and money-effort. Although we did not focus on these parameters in our work, we can provide initial findings. In general, method based on digital preservation is more time consuming at the stages of sampling and sample preservation, regardless of sampling procedure. It is mainly due to the extra time spent on sample preparation before digital preservation. However, processing of samples preserved in alcohol, from bringing it to laboratory until obtaining taxa list, takes more time than digital image analysis. In our case, it was only up to 3 h for single image. Consequently, digitalization might reduce costs even if traditional procedure allows for processing just a subsample. Furthermore, in case of traditional procedures, there is need to buy preserving liquids and maintain laboratory for animal sorting and identification. Due to the fact that sales market offers a wide range of stereoscopic microscope and photographic cameras in terms of quality and price, costs concerning both devices are similar. Moreover, price/quality ratio tends to be reducing. However, as samples preserved in alcohol take space in warehouse, storage of digital images is less expensive. We should also expect prices for digital data storage devices to decrease, while expecting the opposite for storage areas. It is also noteworthy, digital preservation makes sending samples between institutions responsible for bioassessment easier (e.g., for verification of taxa identification).
The results presented above confirm our assumption that assessments of the ecological status of waters based on stream macrobenthos can be made with sufficient accuracy without the killing of necessitated animals. We are thus convinced of the need to further develop the proposed procedure.
References
Álvarez - Cabria, M., J. Barquin & A. Juanes, 2011. Microdistribution patterns of macroinvertebrate communities upstream and downstream of organic effluents. Water Research 45: 1501–1511.
AQEM Consortium, 2002. Manual for the application of the AQEM system. A comprehensive method to assess European streams using benthic macroinvertebrates, developed for the purpose of the Water Framework Directive. Version 1.0, February 2002.
Armitage, P. D., D. Moss, J. F. Wright & M. T. Furse, 1983. The performance of a new biological water quality score system based on macroinvertebrates over a wide range of unpolluted running-water sites. Water Research 17: 333–347.
Bailey, R. C., R. H. Norris & T. B. Reynoldson, 2004. Bioassessment of freshwater ecosystems using the reference condition approach. Kluwer Academic Publishers, Boston: 170.
Barbour, M. T., J. Gerritsen, B. D. Snyder & J. B. Stribling, 1999. Rapid Bioassessment Protocols for Use in Streams and Wadeable Rivers: Periphyton, Benthic Macroinvertebrates and Fish, 2nd ed. U.S. Environmental Protection Agency; Office of Water, Washington D.C.
Birk, S., J. Strackbein & D. Hering, 2010. WISER methods database. Version: March 2011. <http://www.wiser.eu/results/method-database/>.
Bis, B. & A. Mikulec, 2013. Przewodnik do oceny stanu ekologicznego rzek na podstawie makrobezkręgowców bentosowych. [The outline of the ecological status assessment of rivers in Poland, based on the benthic macroinvertebrates assemblages]. Biblioteka Monitoringu Środowiska, Warszawa, pp. 127.
Bonada, N., N. Prat, V. H. Resh & B. Statzner, 2006. Developments in aquatic insect biomonitoring: a comparative analysis of recent approaches. Annual Review of Entomology 51: 495–523.
Cairns Jr., J. & J. R. Pratt, 1993. A history of biological monitoring using benthic macroinvertebrates. In Rosenberg, D. M. & V. H. Resh (eds), Freshwater Biomonitoring and Benthic Macroinvertebrates. Chapman and Hall, New York: 10–27.
Czerniawska-Kusza, I., 2004. Use of artificial substrates for sampling benthic macroinvertebrates in the assessment of water quality of large lowland rivers. Polish Journal of Environmental Studies 13: 579–584.
Davies, P. E., 2000. Development of a national river bioassessment system (AUSRIVAS). In Wright, J. F., D. W. Sutcliffe & M. T. Furse (eds), Assessing the biological quality of fresh waters: RIVPACS and other techniques. Freshwater Biological Association, Ambleside: 113–124.
De Pauw, N. & G. Vanhooren, 1983. Method for biological quality assessment of watercourses in Belgium. Hydrobiologia 100: 153–168.
De Pauw, N., D. Roels & A. P. Fontoura, 1986. Use of artificial substrates for standardized sampling of macroinvertebrates in the assessment of water quality by the Belgian Biotic Index. Hydrobiologia 133: 237–258.
Directive 2000/60/EC. Directive 2000/60/EC of the European Parliament and of the Council of 23 October 2000 Establishing a Framework for Community Action in the Field of Water Policy.
Fleituch, T., H. Soszka, D. Kudelska & A. Kownacki, 2002. Macroinvertebrates as indicators of water quality in rivers: a scientific basis for Polish standard method. Archiv für Hydrobiologie Supplement 141: 225–239.
Hilsenhoff, W., 1988. Rapid field assessment of organic pollution with a family-level biotic index. Journal of the North American Benthological Society 7: 65–68.
Jones, C. F., 2008. Taxonomic sufficiency: the influence of taxonomic resolution on freshwater bioassessments using benthic macroinvertebrates. Environmental Reviews 16: 45–69.
Keizer-Vlek, H. E., P. W. Goedhart & P. F. M. Verdonschot, 2011. Comparison of bioassessment results and costs between preserved and unpreserved macroinvertebrate samples from streams. Environmental Monitoring and Assessment 175: 613–621.
Kołodziejczyk, A. & P. Koperski, 2000. Bezkrezgowce słodkowodne Polski. Klucz do oznaczania oraz podstawy biologii i ekologii makrofauny. [Freshwater macroinvertebrates of Poland. A key to taxa recognition with basis of biology and ecology of macroinvertebrates.] Wydawnictwa Uniwersytetu Warszawskiego, Warszawa: 250 pp.
Kondracki, J., 2002. Geografia regionalna Polski. [Regional geography of Poland]. Wydawnictwo Naukowe PWN, Warszawa: 444 pp.
Koperski, P., 2011. Diversity of freshwater macrobenthos and its use in biological assessment: a critical review of current applications. Environmental Reviews 19: 16–31.
Koperski, P., 2014. The effect of a recognizer – the importance of individual variability and personal ability in animal identification for ecological quality assessment. 2nd International Conference Water Resources and Wetlands. Tulcea, Romania, September 11–13, 2014.
Koperski, P. & M. Gołub, 2006. Application of new regional biotic index apodemac, in environmental quality assessment of lowland streams. Polish Journal of Ecology 54: 311–320.
Kornijow, R. & G. Lechowska, 2002. Effect of treated sewage on benthic invertebrate communities in the upland Bystrzyca Lubelska river. In Kownacki, A., H. Soszka, T. Fleituch & D. Kudelska (eds), River biomonitoring and benthic invertebrate communities. Institute of Environmental Protection, Warszawa: 45–52.
Kownacki, A. & H. Soszka, 2004. Wytyczne do oceny stanu rzek na podstawie makrobezkręgowców oraz do pobierania prób makrobezkręgowców w jeziorach [Guidelines for the assessment of rivers based on macroinvertebrates and sampling macroinvertebrates in lakes]. Zakład Ochrony Przyrody PAN, Warszawa: 54 pp.
Kownacki, A., T. Fleituch & E. Dumnicka, 2002. The effect of treated wastes on benthic invertebrate communities in the mountain zone of the Dunajec river (southern Poland). In Kownacki, A., H. Soszka, T. Fleituch & D. Kudelska (eds), River biomonitoring and benthic invertebrate communities. Institute of Environmental Protection, Warszawa: 22–49.
Lenat, D. R., 1988. Water quality assessment of streams using a qualitative collection method for benthic macroivertebrates. Journal of North American Bethological Society 7: 222–233.
Norris, R. H., 1995. Biologcal monitoring: the dilemma of data analysis. Journal of North American Bentological Society 14: 440–450.
Odum, E. P., J. T. Finn & E. H. Franz, 1979. Perturbation theory and the subsidy-stress gradient. BioScience 29: 349–352.
Orlofske, J. M. & D. J. Baird, 2013. The tiny mayfly in the room: implications of size-dependent invertebrate taxonomic identification for biomonitoring data properties. Aquatic Ecology 47: 481–494.
Pfrender, M. E., C. P. Hawkins, M. Bagley, G. W. Courtney, B. R. Creutzburg, J. H. Epler, S. Fend, D. Schindel, L. C. Jr Ferrington, P. L. Hartzell, S. Jackson, D. P. Larsen, A. Lévesque, J. C. Morse, M. J. Petersen, D. Ruiter & M. Whiting, 2010. Assessing macroinvertebrate biodiversity in freshwater ecosystems: advances and challenges in DNA-based approaches. Quarterly Review of Biology 85: 319–340.
Plafkin, J. L., M. T. Barbour, K. D. Porter, S. K. Gross & R. M. Hughes, 1989. Rapid bioassessment protocols for use in streams and rivers: benthic macroinvertebrates and fish. U.S. Environmental Protection Agency, Office of Water Regulations and Standards, Washington, DC.
Schneider, C. A., W. S. Rasband & K. W. Eliceiri, 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9: 671–675.
Shannon, C. E., 1948. A mathematical theory of communication. The Bell System Technical Journal 27(379–423): 623–656.
StatSoft, Inc. (2011). STATISTICA (data analysis software system), version 10. www.statsoft.com.
Smith, M. J., W. R. Kay, D. H. D. Edward, P. J. Papas, K. St, J. Richardson, J. C. Simpson, A. M. Pinder, D. J. Cale, P. H. J. Horwitz, J. A. Davis, F. H. Yung, R. H. Norris & S. A. Halse, 1999. AUSRIVAS: using macroinvertebrates to assess ecological condition of rivers in Western Australia. Freshwater Biology 41: 269–282.
Sweeney, B. W., J. M. Battle, J. K. Jackson & T. Dapkey, 2011. Can DNA barcodes of stream macroinvertebrates improve descriptions of community structure and water quality? Journal of the North American Benthological Society 30(1): 195–216.
Verdonschot, P. F. M., 2006. Data composition and taxonomic resolution in macroinvertebrate stream typology. Hydrobiologia 566: 59–74.
Acknowledgments
We are very grateful to two anonymous reviewers for many valuable comments that helped to improve previous version of the manuscript. The study was carried out at the Biological and Chemical Research Centre, University of Warsaw, established within the project co-financed by European Union from the European Regional Development Fund under the Operational Programme Innovative Economy, 2007–2013. The financial support has been provided by grant of National Science Centre of Republic of Poland No. 2011/01/B/NZ9/02590.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling editor: Sonja Stendera
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Karasek, T., Koperski, P. NoMBSI: a new, Non-lethal Method for Benthos Sampling and Identification for use in biological monitoring of flowing waters: preliminary results. Hydrobiologia 751, 215–227 (2015). https://doi.org/10.1007/s10750-015-2188-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-015-2188-2