Abstract
Nuclear factor I-C (NFIC) plays critical roles in the regulation of tooth development by influencing the biological behaviors of stem cells in the dental germ. This study aimed to investigate the effect of NFIC on the vitality and osteogenic/cementogenic differentiation of rat dental follicle cells (DFCs). DFCs were isolated from dental follicles in the first molars of neonatal rats. DFCs expressed mesenchymal stromal cell markers CD29, CD44 and CD90 and had capabilities for self-renewal and multipotent differentiation. Overexpression of NFIC promoted the proliferation of DFCs without markedly influencing the apoptosis of DFCs. Moreover, NFIC increased alkaline phosphatase (ALP) activity in DFCs and upregulated the mRNA levels of osteogenic-related markers, namely, collagen type I (Col I), Runt-related transcription factor 2 (Runx2) and ALP, as well as β-catenin. In contrast, silencing NFIC by siRNA increased the apoptosis of DFCs and downregulated the expression of osteogenic-related markers. In conclusion, these results suggested that upregulation of NFIC may promote the proliferation and osteogenic/cementogenic differentiation of DFCs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Periodontal disease, which is one of most common oral diseases in adults, destroys the periodontal apparatus, including tooth-supporting alveolar bones (ABs), and results in tooth loosening and even tooth loss. Many researchers have focused on the regeneration of periodontal tissues; however, the cellular and molecular bases of periodontal tissue formation and repair are unclear, and clinical periodontal regeneration has still not been successfully developed to meet the need despite decades of research efforts in this area (Pan et al. 2010). According to the current knowledge of periodontal development, the dental follicle is the origin of periodontal tissues, which is an ectomesenchymal tissue that surrounds the tooth germ and plays a critical role in periodontal development and tooth eruption (Chalisserry et al. 2017; Wise et al. 1985). Undifferentiated ectomesenchymal cells, which are known as dental follicle stem cells or dental follicle cells (DFCs), can differentiate and form cementum, periodontal ligament (PDL) and AB during the process of periodontal tissue development (Yokoi et al. 2007; Sowmya et al. 2015; Guo et al. 2012a, b; Shuai et al. 2018). Root-like tissues formed in the alveolar fossa with positive staining for markers of periodontal tissues and dental pulp when DFCs were transplanted into a treated dentin matrix scaffold (Guo et al. 2012a, b). Interestingly, DFCs rescue the degeneration of inflamed periodontal ligament stem cells (PDLSCs), suggesting that DFCs may aid PDLSCs in regenerating periodontal tissues by ameliorating the local microenvironment (Liu et al. 2014). Furthermore, after cryopreservation, human DFCs have been demonstrated to be a reliable resource for regenerative medicine (Park et al. 2017). Due to their outstanding multiple differentiation competencies, DFCs have been selected as promising stem cells for cell therapy in terms of periodontal regeneration. However, a better understanding of the properties of DFCs and their application in periodontal regeneration is still necessary.
The nuclear factor I (NFI) family of transcription–replication factors consists of four genes (NFIA, NFIB, NFIC, and NFIX) in mammals (Gronostajski 2000; Rupp et al. 1990). NFIC is regarded as a key regulator of tooth development. NFIC deficiency causes aberrant odontoblasts and abnormal dentin, as well as periodontium formation, which ultimately lead to short molar roots (Park et al. 2007). Accumulative evidence has revealed that NFIC is involved in odontogenic cell proliferation, differentiation and survival during root formation (Lee et al. 2009a, b; Oh et al. 2012). Strong NFIC expression has been detected in the dental papilla and dental follicle during molar development in rats (Chen et al. 2014). Considering the importance of NFIC in root development and the potential osteoblast differentiation of DFCs, gene-modified seed cells can provide continued strategies for the overexpression of specific transgenes and broaden the therapeutic capabilities for tissue engineering and regeneration (Yu et al. 2018; Hu et al. 2018; Tan et al. 2009). Thus, we hypothesized that NFIC influences the biological properties (proliferation, apoptosis and osteogenesis) of DFCs and regulates the osteogenic/cementogenic differentiation of DFCs, which maybe candidate stem cells for regenerative therapy.
Materials and methods
Cell cultures
All experiments were performed with the approval of the Ethics Committee of Guanghua School and Hospital of Stomatology, Sun Yat-sen University, China. Sprague–Dawley rats were purchased from the animal research center at Sun Yat-sen University. Rats were sacrificed at postnatal (PN) days 1, 3, 5, and 7 by an overdose of 10% chloralhydrate solution. Dental follicle tissues were carefully isolated from the first molar using a dental explorer, minced into 1 × 1 × 1-mm3 pieces, plated in 60-mm culture dishes, and then cultured in α-MEM medium (Gibco, Grand Island, New York, USA) supplemented with 20% FBS (Gibco, Grand Island, New York, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37 °C in a 5% CO2 incubator. Cells were digested using 0.25% trypsin and 0.25 mM EDTA (Gibco, Grand Island, New York, USA) and passaged at a ratio of 1:2 when the cells reached approximately 80–90% confluence. DFCs at passages 3–5 were used in subsequent analyses. For osteogenic induction, the cells were cultured in osteogenic (OS) medium containing 10% FBS, 200 μM ascorbic acid, 10 mM β-glycerophosphate and 100 nM dexamethasone (Sigma-Aldrich, St. Louis, MO, USA).
Immunocytochemical staining
DFCs were fixed in freshly prepared 4% paraformaldehyde (pFA) and permeabilized with 0.3% Triton X-100, endogenous peroxidase was removed with 0.3% H2O2 for 30 min, and DFCs were incubated with 5% goat serum for 30 min at 37 °C. Mouse anti-rat cytokeratin or mouse anti-rat vimentin antibodies (BOSTER, China) were used as primary antibodies at dilutions of 1:100 for 2 h at 37 °C. The cells were developed in 3,3′-diaminobenzidine (DAB) for 60–90 s and counterstained with hematoxylin for 5 min. Images were captured with an inverted microscope (Axio Observer Z1; Carl Zeiss AG, Oberkochen, Germany).
Osteogenic/adipogenic induction
When cells reached approximately 70–80% confluence, the medium was replaced with OS or adipogenic medium (1 μM dexamethasone, 200 μM indomethacin, and 500 μM 3-isobutyl-1- methylxanthine; Sigma-Aldrich, St. Louis, MO, USA) for 21 or 14 days. The cells were stained with 1% alizarin red S solution (Cyagen, China) or 3 mg/mL oil red O (Sigma-Aldrich, St. Louis, MO, USA). Images were captured with an inverted microscope.
Colony-formation assay
A colony-formation assay was performed to determine the proliferative potential of DFCs. Cells were seeded in 10-cm culture dishes at a low density (100 cells/plate) and cultured in α-MEM medium supplemented with 10% FBS and 100 U/mL penicillin–streptomycin for 14 days. Subsequently, the colonies were stained with 0.1% crystal violet for 20 min. The number of colonies with > 50 cells was counted under an inverted microscope. Colony-formation efficiency was calculated by dividing the total number of colonies by the number of seeded cells.
Flow cytometric analysis of DFCs
DFCs were prepared as single-cell suspensions in α-MEM medium supplemented with 2% FBS. Cells were incubated with CD29, CD34, CD44, CD45 and CD90 antibodies (BD Biosciences, San Diego, CA, USA) for 30 min at 4 °C in the dark. Samples were analyzed by flow cytometry (Beckman-Coulter, CA, USA).
Western blotting and immunocytochemical staining for NFIC in DFCs
Total protein in DFCs at different developmental stages (PN days 1–7) was extracted using RIPA lysis buffer, and the protein concentration was determined using a Bicinchoninic Acid protein kit (Beyotime, Shanghai, China) according to the manufacturer’s protocol. Protein lysates (40 µg) were separated using 12% SDS-PAGE gels and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA, USA). The membranes were blocked with 5% skim milk at room temperature for 2 h and incubated with monoclonal mouse anti-rat NFIC antibody (1:1000, Abcam, Burlingame, CA, USA) and monoclonal mouse anti-rat β-actin (1:1000, Beyotime, Shanghai, China) overnight at 4 °C. β-actin was used as an internal control. The secondary antibody was horseradish peroxidase-conjugated goat anti-mouse IgG secondary antibody (1:1000, Beyotime, Shanghai, China). Immunoreactive bands were visualized using an ECL-Westernblot kit (CWBiotech, China). The densitometry of the bands was quantified with ImageJ 1.36b (NIH Freeware, USA). Immunocytochemical staining for NFIC in DFCs at PN day 5 was performed according to the protocol described for immunocytochemical staining for cytokeratin and vimentin.
Lentiviral transduction
siNFIC sense lentivirus and control lentivirus were obtained and synthesized by Hanbio (Shanghai, China). A lentivirus to overexpress rat NFIC and control plasmid DNA was designed and constructed (Hanbio, Shanghai, China). DFCs were infected with lentivirus (MOI = 40) at 50–60% confluence in the presence of 4 μg/mL Polybrene. Then, the cells were cultured in media containing 2 μg/mL puromycin. Real-time PCR and Western blotting were used to analyze the mRNA and protein levels, respectively, of NFIC.
Real-time quantitative PCR analysis
Total cellular mRNA was isolated using TRIzol (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s protocol. cDNA was obtained using a Transcriptor First Strand cDNA Synthesis kit (Roche Applied Science, Penzberg, Bayern, Germany). The reverse transcription product was subjected to real-time PCR using SYBR Green I Master (Roche Applied Science, Penzberg, Bayern, Germany). The thermocycling conditions consisted of 95 °C for 5 min; 45 cycles at 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 20 s; 1 cycle at 95 °C for 5 s, 65 °C for 1 min, and 97 °C; and a final 10-s cooling step at 40 °C. The PCR primer sequences are provided in Table 1. PCR products were analyzed on a 2% agarose gel.
Cell counting Kit-8 assay
DFCs were seeded at a density of 2000 cells/well in 96-well plates and maintained in α-MEM medium supplemented with 10% FBS and 100 U/mL penicillin–streptomycin. At specific time points (1, 2, 3, 4, 5, 6, and 7 days), the cells were incubated with 10% Cell Counting Kit-8 (CCK8, Dojindo, Japan) solution for 2 hat 37 °C. Cell viability was analyzed by measuring the absorbance at 450 nm in an ELISA plate reader (Tecan, Grodig, Austria). Each group was analyzed in three parallel wells.
Cell cycle analysis
After lentiviral transduction, DFCs were prepared as single-cell suspensions, washed with cold PBS and then fixed in 70% ethanol at 4 °C overnight. The cells were washed with cold PBS twice and incubated in 0.5 mL of PI/RNase Staining Solution (Beyotime, Shanghai, China) for 30 min at 37 °C. Cell cycle analysis of 10,000 cells per sample was carried out using a CytoFLEX flow cytometer (Beckman-Coulter, CA, USA).The data were analyzed with CytExpert analysis 2.0 software.
TUNEL assay
After lentiviral transduction, DFCs were seeded at a density of 5000 cells/well in 48-well plates and maintained in α-MEM medium supplemented with 10% FBS and 100 U/mL penicillin–streptomycin for 24 h. Cell apoptosis was examined with a terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay kit (Roche Applied Science, Penzberg, Bayern, Germany) and by DAB staining (Beyotime, Shanghai, China) according to the manufacturer’s protocol. The index of apoptosis was calculated as the percentage of apoptotic (TUNEL-positive) cells to the total (DAPI-positive) number of cells using Image-Pro Plus Software (Rockville, MD, USA).
Flow cytometry analysis of apoptosis
After lentiviral transduction, DFCs were prepared as single-cell suspensions, washed with cold PBS, and incubated with Annexin V-APC and PI (Elabscience Biotechnology, Wuhan, China) for 15 min at room temperature. Cell samples were analyzed and isolated using a CytoFLEX flow cytometer (Beckman-Coulter, CA, USA).
Alkaline phosphatase (ALP) activity assay
Cells were treated for osteogenic induction for 3 days, and total protein was extracted using RIPA lysis buffer. Protein samples were stained with 0.5 mg/mL p-nitrophenyl phosphate from a commercial ALP kit (Jiancheng, Nanjing, China). The activity of ALP was determined by measuring the absorbance at 520 nm using an ELISA plate reader (Tecan, Grodig, Austria) and normalized to the protein concentration.
Statistical analysis
Data from each assay in this study were obtained from at least three independent experiments. Experimental quantitative values are presented as the mean ± standard deviation (SD). The two-tailed Student’s t test (SPSS 19.00) was used for the two-subgroup comparisons. One-way ANOVA (SPSS 19.00) was used to compare the means between multiple groups, and the least significant difference (LSD) was used for post hoc analysis. P < 0.05 was considered indicative of statistical significance.
Results
Isolation and identification of DFCs
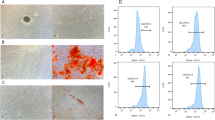
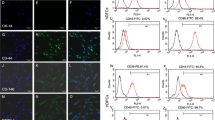
Small pieces of dental follicle tissue were cultured in a 25-cm2 flask from which DFCs grew following 2 days of culture (Fig. 1A(a)). DFCs showed a fibroblast-like morphology with centrally localized single nuclei and abundant cytoplasm. Cells were digested, passaged and then used for further studies when they grew to near-complete confluence (Fig. 1A(b)). The DFCs in this study were vimentin positive (Fig. 1B(a)) and cytokeratin negative (Fig. 1B (b)) according to immunocytochemistry staining, which was consistent with the properties of stromal cells. After osteogenic or adipogenic induction, DFCs were found to form mineralized nodules (Fig. 1C(a)) or lipid droplets (Fig. 1C(b)), respectively. After 14 days of colony-formation culture, DFCs formed colonies (Fig. 1D(a)) and were arranged in clusters (Fig. 1D(b)). The colony-formation efficiency was ~ 24% (± 5.2%; n = 3). The expression levels of mesenchymal markers, including CD29 (98.67%), CD44 (84.73%), and CD90 (99.98%), were high (Fig. 1E), while the expression levels of hematopoietic markers CD34 and CD45 were very low in DFCs according to flow cytometric analysis (Fig. 1E).
Characterization of rat dental follicle cells (DFCs). A Primary culture: DFCs were migrated from a dental follicle tissue block after 2 days of culture (a), and DFCs grew to near confluence after 7 days (b). B Immunocytochemical staining reveals that DFCs are positive for vimentin (a) and negative for cytokeratin (b). C After being separately cultured in osteogenic (OS) and adipogenic media, mineralized nodules were stained with alizarin red (a), and oil droplets were stained with oil red O (b). D Colony-formation assay: gross appearance (a) and microscopic appearance (b). E Flow cytometric analysis reveals that DFCs are positive for CD29, CD44 and CD90 but negative for CD34 and CD45. Scale bar 100 μm
NFIC expression in DFCs
NFIC expression was examined by Western blotting at PN days 1, 3, 5, and 7 to investigate its expression pattern during different developmental stages of the tooth. The protein expression of NFIC was increased in a time-dependent manner, and maximum expression (threefold increase) was observed at PN day 5 or 7 (Fig. 2A). Immunocytochemical staining for NFIC showed positive staining in DFCs at PN day 5 (Fig. 2B(b)), and the negative controls are shown in Fig. 2B(a).
Protein levels of NFIC in DFCs. A Changes in NFIC protein expression in DFCs at PN days 1, 3, 5, and 7 were determined by Western blotting. Representative immunoblots are shown on the left, and the values were normalized to β-actin. The optical density was measured, and each value is shown as the mean ± SD (right); n = 3. (*p < 0.05). B Immunocytochemistry for NFIC in DFCs at PN day 5 (b) and the negative control (a) are shown. Scale bar 100 μm
Knockdown and overexpression of NFIC by lentiviral transduction
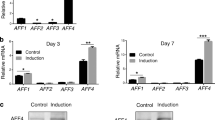
To determine the importance of NFIC in DFCs, we employed a lentiviral transduction approach to knock down or overexpress NFIC. DFCs were transduced with the interference lentivirus siNFIC (LV siNFIC), interference lentivirus siCtrl (LV siCtrl), lentivirus that expressed NFIC (LV NFIC), and lentivirus containing control plasmid DNA (LV Ctrl), and the DFCs were all positive for GFP (Fig. 3A).Quantitative analysis revealed an 82.3% (± 3.9; n = 3, p < 0.01) and a 76.7% (± 7.1; n = 3, p < 0.001) decrease in the NFIC mRNA (Fig. 3B) and protein levels (Fig. 3C), respectively, compared to those of the scrambled siRNA (Fig. 3B). Quantitative analysis revealed that the LV NFIC cells had 30.3-fold (± 16.6; n = 3, p < 0.05) and 6.7-fold (± 3.2; n = 3, p < 0.001) higher NFIC mRNA (Fig. 3D) and protein levels (Fig. 3E), respectively, than the LV-vector control cells.
Knockdown and overexpression of NFIC by lentiviral transduction. A DFCs were transduced with an RNA interference lentiviral control vector (lentivirus sicontrol, LV siCtrl), NFIC RNA interference lentiviral vector (lentivirus siNFIC, LV siNFIC), control plasmid DNA vector (lentivirus control, LV Ctrl) or a plasmid containing DNA encoding NFIC (LV NFIC). The efficiency of lentivirus transduction was determined by examining GFP-positive cells under a microscope. B The efficiency of NFIC knockdown after infection with lentivirus was analyzed by qRT-PCR in LV siCtrl or LV siNFIC cells. **p < 0.01 versus LV siCtrl. C The efficiency of NFIC knockdown was analyzed by Western blotting in LV siCtrl or LV siNFIC cells (left panel). The values were normalized to β-actin. ***p < 0.001 versus LVsiCtrl. D The efficiency of NFIC overexpression after infection with lentivirus was analyzed by qRT-PCR in LV Ctrl or LV NFIC cells. *p < 0.05 versus LV Ctrl. e The efficiency of NFIC overexpression was analyzed by Western blotting in LV Ctrl or LV NFIC cells (left panel). The values were normalized to β-actin. ***p < 0.001 versus LV Ctrl. Data are the mean ± SD of three independent experiments. Scale bar 100 μm
Effects of NFIC on the proliferation and apoptosis of DFCs
Next, we explored the effect of NFIC knockdown or overexpression on the proliferation of DFCs. As shown in Fig. 4A, the proliferation rate of LV NFIC cells was higher than that of LV Ctrl cells or control cells. All types of cells proliferated slowly from 1 to 2 days. However, from 3 to 5 days, LV siNFIC cells grew more slowly than LV siCtrl cells (Fig. 4A, n = 5, p < 0.05 or p < 0.01).Cell cycle analysis showed that NFIC knockdown delayed cell cycle progression by arresting cells in the G0/G1 phase, and that NFIC overexpression increased cell proliferation by upregulating cells from the G0/G1 phase to the S phase (Fig. 4B, C, n = 3).
Effects of NFIC knockdown or over expression on the proliferation of DFCs. A Cell viability was measured by CCK-8 assay. Data are expressed as the mean ± SD of five independent experiments. *p < 0.05, **p < 0.01 versus LV siCtrl or LV Ctrl, respectively. B Cellular DNA contents were then examined using flow cytometry. C The relative G0/G1 phase fraction (%) in cell cycle progression is presented. Statistical measures were expressed as the mean ± SD of three independent experiments. ***p < 0.001 versus LV siCtrl, *p < 0.05 versus LV Ctrl
The apoptotic effect of NFIC was examined by TUNEL staining, which showed the occurrence of chromatin condensation and nuclear fragmentation. In the control groups, including control cells, LV siCtrl cells and LV Ctrl cells, most of the cells appeared well attached, some cell division was apparent (black arrow, Fig. 5A upper panel), and few cells were undergoing apoptosis (red arrow, Fig. 5A upper panel). Knockdown of NFIC increased the percentages of apoptotic cells to 18.16 ± 3.04% from 1.77 ± 0.36% under LV siCtrl conditions (Fig. 5B, n = 3, p < 0.001). In contrast, overexpression of NFIC did not affect the apoptosis of DFCs, and more cell division was apparent. More evident results were obtained by flow cytometry analysis (Fig. 5A lower panel and c, n = 3, p < 0.01).
Effects of NFIC knockdown or over expression on cell apoptosis of DFCs. A Apoptosis was determined by TUNEL staining (upper panel) and flow cytometric analysis (lower panel). Red arrows indicate apoptotic cells, and black arrows indicate cell division. B, C The apoptotic rate determined by TUNEL staining (B) and flow cytometric analysis (C) was calculated. Data are expressed as the mean ± SD of three independent experiments. ***p < 0.001 versus LV siCtrl; **p < 0.01 versus LV siCtrl; Scale bar 100 μm
Effects of NFIC on the osteogenesis of DFCs
The osteogenic potential of DFCs was measured by assessing ALP activity and performing qRT-PCR for the gene expression of Runt-related transcription factor 2 (Runx2), collagen type I (Col I), and ALP. As shown in Fig. 6A, B, the ALP activity increased when DFCs were exposed to OS medium. ALP activity increased significantly (Fig. 6B, n = 4, p < 0.001) in LV NFIC cells supplemented with OS medium (OS+) compared with that in LV Ctrl cells exposed to OS medium. In contrast, ALP activity decreased significantly (Fig. 6A, n = 4, p < 0.05) in LV siNFIC cells versus LV siCtrl cells when they were exposed to OS medium. Similarly, supplementation of LV NFIC cells with OS medium (OS+) resulted in significant increases in the gene expression of ALP, Runx2 and Col I, which are markers of osteogenesis (Fig. 6D). Knockdown of NFIC decreased the gene expression of Col I, Runx2 and ALP (Fig. 6C). Interestingly, the level of β-catenin was increased significantly in LV NFIC cells supplemented with osteogenic medium (OS+). The expression of β-catenin was consistent with the change in the osteogenic potential of DFCs (Fig. 6E, F).
Effects of NFIC knockdown or overexpression on the osteogenic differentiation of DFCs. A, B After osteogenic induction for 3 days, ALP activity in the lysates of DFCs infected with lentivirus was measured by colorimetric assay. Each value is presented as the mean ± SD; n = 4. C, D The mRNA levels of ALP, Runx2 and Col I were analyzed by qRT-PCR. Data are expressed as the mean ± SD; n = 4. E, F The mRNA levels of β-catenin were analyzed by qRT-PCR. Data are expressed as the mean ± SD; n = 3. *p < 0.05, **p < 0.01, ***p < 0.001 versus Ctrl cells without osteogenic induction (OS−).#p < 0.05, ##p < 0.01, ###p < 0.001 versus LV siCtrl or LV Ctrl cells with osteogenic induction (OS+)
Discussion
Periodontitis, a chronic inflammatory disease, results in disruption of periodontal tissues and progressive AB destruction. Stem-cell-based tissue engineering has been increasingly emphasized as a novel therapeutic strategy for periodontal repair. To date, many advances have been achieved in stem cell-based tooth and periodontal regeneration (Hu et al. 2018). However, in periodontitis, varying degrees of damage to various types of tissues, such as the PDL, cementum, or AB, and complex tissues are involved. The optimal intervention strategy for these conditions is to restore all of the heterogeneously affected tissues (Guo et al. 2012a, b). Correspondingly, DFCs are the precursor cells of PDL cells, osteoblasts, and cementoblasts, which lead to the formation of the PDL, cementum, and AB, respectively. Therefore, DFCs have the potential to be one of most suitable seeding cell types for regeneration of heterogeneous periodontal tissue.
DFCs were first isolated from the dental follicle of the human third molar (Morsczeck et al. 2005). DFCs can also be obtained from different species, including mice and rats (Yao et al. 2008; Luan et al. 2006). Considering the ethical dilemmas and issues of convenience involved in DFC isolation, DFCs acquired from the first molar dental follicle of rats have the advantages of being easier to harvest, providing a sufficient tissue block of the dental follicle, and being subsequently and successfully cultured in vitro, reflecting potential cost-effective and efficient approach (Yao et al. 2008). DFCs met the criteria for being MSCs, which can differentiate into a variety of cell types, exhibit plastic adherent fibroblast-like cell growth and positively express specific stromal markers (Dominici et al. 2006). In this study, DFCs were acquired from dental follicles and found to be adherent fibroblastic cells with positivity for stromal markers CD29, CD44 and CD90 and negativity for hematopoietic markers CD34 and CD45. DFCs also showed the capacity of self-renewal and osteogenic and adipogenic differentiation. These results demonstrated that DFCs were stromal in origin and had MSC features without hematopoietic contamination, which confirms the results of previous studies (Pan et al. 2010; Yao et al. 2008).
During tooth development, NFIC has been reported to first be detected in the crown of the first mandibular molar when hard tissue starts to form at PN day 4 and is observed in DFCs, DPCs and crown ameloblasts (Chen et al. 2014). Thus, our team isolated DFCs from the first molars of PN day 1 to 7 rats to investigate the expression pattern of NFIC. The results showed that NFIC protein expression was increased in a time-dependent manner, and maximum expression was observed at PN day 5 or 7, implying that NFIC may have specific effects on DFC behavior.
Sufficient and high numbers of proliferating cells are required in periodontal regeneration to compensate for tissue defects. We demonstrated that NFIC can markedly promote the proliferation of DFCs, which is consistent with the findings of Lee et al. (Lee et al. 2009a, b) who revealed the proliferative and antiapoptotic activities of NFIC in odontogenic cells. Moreover, overexpression of NFIC increases cell proliferation in SCAPs (Zhang et al. 2015), while NFIC silencing delayed the proliferation of DFCs in our study. In addition, NFIC-deficient primary pulp cells show decreased proliferation activity in vitro (Lee et al. 2009a, b). Zhou also reported that NFIC silencing inhibits cell proliferation in progenitor cells (Zhou et al. 2017). Nfic gene inactivation has been reported to lead to an increase in the expression of cell cycle inhibitors such as p21 and p16, which is consistent with a recent finding that NFI is a key repressor of p21 transcription (Lee et al. 2009a, b). Disabling of NFIC has been demonstrated to enhance the expression of cell cycle inhibitors p16 and p21 but suppress the expression of cyclin B1 and cyclin D1 in pulp cells, leading to suppression of proliferation in odontogenic cells (Lee et al. 2009a, b). Similarly, progenitor cell proliferation and hair follicle regeneration are delayed without NFIC, implying impaired activation of the hair follicle cycle (Plasari et al. 2010). Likewise, NFIC silencing prolongs the G1 phase of the cell cycle in the apical papilla (hSCAPs) (Gao et al. 2014). NFIC silencing results in G1-phase arrest of hSCAPs, indicating that NFIC may participate in the regulation of cell cycle progression. In the present study, an overexpression NFIC plasmid was constructed into a lentivirus and transduced into DFCs to obtain cells that overexpressed NFIC. Overexpression of NFIC promoted the proliferation of DFCs, while transduction of an empty vector had a minimal effect on the proliferation of DFCs, indicating that the presence of the lentivirus had little influence on cell growth.
Apoptosis is an essential physiological process that plays an important role in periodontal repair. Our study showed that the apoptosis of DFCs was not affected by NFIC overexpression, and that more cell division was apparent in this setting. Knockdown of NFIC increased the number of apoptotic DFCs. These results are similar to those in Zhou’s report indicating that NFIC silencing promotes the apoptosis of progenitor ST2 cells (Zhou et al. 2017). In addition, Nfic deficiency is responsible for odontoblast apoptosis, and odontogenic cells in Nfic-deficient mice may undergo apoptosis via Smad-dependent pathways (Lee et al. 2009a, b).
To investigate the role of NFIC in osteoblastic differentiation of DFCs, we assessed the osteogenesis capacity of DFCs after NFIC activation. Col I, ALP and Runx2 are frequently selected as specific markers to detect the osteogenic or cementogenic potential of dental MSCs (Park et al. 2012; Lee et al. 2012; Ercal et al. 2018). ALP activity is increased in the early stages of osteoblast commitment. During osteogenic differentiation, upregulation of ALP activity is assumed to reflect the number of osteogenic-committed progenitor cells in a cell population. Induction of ALP activity may serve as a predictive parameter for in vivo bone formation (Prins et al. 2014). Runx2, a master transcription factor, is essential for tooth and skeletal development and plays an important role in the multilineage differentiation of DFCs (Pan et al. 2010). Our data showed that NFIC significantly promoted ALP activities in DFCs. Enhanced ALP activity was consistent with the increased mineralization induced by NFIC. Moreover, NFIC promoted the mRNA expression of several osteogenic markers (Col I, Runx2 and ALP).
To further explore the effect of NFIC on the differentiation of DFCs, NFIC gene silencing was also performed. NFIC-knockdown cells displayed decreases in ALP and Col I expression levels, indicating that the osteoblastic/cementoblastic differentiation of DFCs is suppressed by NFIC inhibition. Similarly, in SCAPs, upregulation of NFIC activity promotes osteo/odontoblastic differentiation of SCAPs (Zhang et al. 2015). Overexpression of NFIC promotes cell proliferation, mineralization and ALP activity in SCAPs. Moreover, NFIC increases the mRNA levels of ALP, osteocalcin and Col I, as well as dentin sialoprotein protein (Dspp) levels, which are considered odontogenic-related markers. On the other hand, knockdown of NFIC by siRNA inhibits the expression of odontogenic-related markers and suppresses the mineralization capacity of SCAPs. Perturbation of NFIC in progenitor cell lines also affects osteoblast differentiation. NFIC overexpression in ST2 cells promotes ALP staining in differentiated osteoblasts and elevates the mRNA and protein levels of osterix, Runx2 and ALP, while NFIC silencing in ST2 cells blunts the mRNA and protein levels of Runx2, osterix and ALP. In apical odontoblasts (AOds), several transcription factors and markers, including Nfic, Osx and Creb3l1, as well as ALP and β-catenin, are expressed. Notably, strong expression of β-catenin is detected in AOds. The expression of ALP and NFIC overlaps in AOds, indicating a possible link between β-catenin and NFIC (Bae et al. 2013).
Interestingly, in the present study, we also found that the expression of β-catenin is consistent with the change in the osteogenic potential of DFCs. β-catenin is both a cell adhesion protein and a transducer within the canonical Wnt/β-catenin pathway, which is a critical molecular signaling pathway during osteogenesis (Bae et al. 2013). A close relationship between NFIC and Wnt signaling may exist. NFIC reciprocally regulates the osteogenic and adipogenic differentiation of progenitor cells. In addition, mechanistic studies of NFIC revealed that NFIC activates canonical Wnt signaling and promotes the nuclear translocation of β-catenin based on the fact that NFIC directly binds to the promoter of Lrp5 and transcriptionally regulates Lrp5 expression (Zhou et al. 2017). Additionally, Wnt5a has been expressed consistently in the rat periodontium from PN day 1 to 11. The proliferation rates of DFCs are significantly reduced after Wnt5a overexpression (Xiang et al. 2014). Constitutive stabilization of β-catenin in the dental mesenchyme results in excessive formation of cementum (Nemoto et al. 2016). Likewise, canonical Wnt signaling can induce DFC differentiation toward a cementoblast/osteoblast phenotype, and Wnt3a increases ALP expression in DFCs. However, the precise mechanisms are still unclear. Further investigations should be conducted to understand the role of NFIC in cementogenesis or osteogenesis, which will lead to the advancement of oral regenerative therapies.
In summary, our results suggested that NFIC enhances the proliferation and osteogenic/cementogenic differentiation of DFCs, as well as the expression of β-catenin, while NFIC gene silencing leads to the apoptosis of DFCs and suppresses the osteoblastic/cementoblastic differentiation of DFCs. Our findings may provide new insights into the molecular mechanism of regulating DFCs to achieve periodontal regeneration and may facilitate the design of new therapeutic interventions for periodontal diseases. However, further animal or in vivo experiments are needed to determine the role of NFIC in periodontal tissue regeneration.
References
Bae CH, Kim TH, Chu JY, Cho ES (2013) New population of odontoblasts responsible for tooth root formation. Gene Expr Patterns 13:197–202. https://doi.org/10.1016/j.gep.2013.04.001
Chalisserry EP, Nam SY, Park SH, Anil S (2017) Therapeutic potential of dental stem cells. J Tissue Eng 8:1545535357. https://doi.org/10.1177/2041731417702531
Chen X, Chen G, Feng L, Jiang Z, Guo W, Yu M, Tian W (2014) Expression of Nfic during root formation in first mandibular molar of rat. J Mol Histol 45:619–626. https://doi.org/10.1007/s10735-014-9588-x
Dominici M et al (2006) Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 8:315–317. https://doi.org/10.1080/14653240600855905
Ercal P, Pekozer GG, Kose GT (2018) Dental stem cells in bone tissue engineering: current overview and challenges. Adv Exp Med Biol 1107:113–127. https://doi.org/10.1007/5584_2018_171
Gao S, Zhao YM, Ge LH (2014) Nuclear factor I-C expression pattern in developing teeth and its important role in odontogenic differentiation of human molar stem cells from the apical papilla. Eur J Oral Sci 122:382–390. https://doi.org/10.1111/eos.12151
Gronostajski RM (2000) Roles of the NFI/CTF gene family in transcription and development. Gene 249:31–45. https://doi.org/10.1016/s0378-1119(00)00140-2
Guo W et al (2012a) Dental follicle cells and treated dentin matrix scaffold for tissue engineering the tooth root. Biomaterials 33:1291–1302. https://doi.org/10.1016/j.biomaterials.2011.09.068
Guo W, Chen L, Gong K, Ding B, Duan Y, Jin Y (2012b) Heterogeneous dental follicle cells and the regeneration of complex periodontal tissues. Tissue Eng Part A 18:459–470. https://doi.org/10.1089/ten.TEA.2011.0261
Hu L, Liu Y, Wang S (2018) Stem cell-based tooth and periodontal regeneration. Oral Dis 24:696–705. https://doi.org/10.1111/odi.12703
Lee DS et al (2009a) Nuclear factor I-C is essential for odontogenic cell proliferation and odontoblast differentiation during tooth root development. J Biol Chem 284:17293–17303. https://doi.org/10.1074/jbc.M109.009084
Lee TY et al (2009b) Disruption of Nfic causes dissociation of odontoblasts by interfering with the formation of intercellular junctions and aberrant odontoblast differentiation. J Histochem Cytochem 57:469–476. https://doi.org/10.1369/jhc.2009.952622
Lee JH, Um S, Jang JH, Seo BM (2012) Effects of VEGF and FGF-2 on proliferation and differentiation of human periodontal ligament stem cells. Cell Tissue Res 348:475–484. https://doi.org/10.1007/s00441-012-1392-x
Liu J, Wang L, Liu W, Li Q, Jin Z, Jin Y (2014) Dental follicle cells rescue the regenerative capacity of periodontal ligament stem cells in an inflammatory microenvironment. PLoS ONE 9:e108752. https://doi.org/10.1371/journal.pone.0108752
Luan X, Ito Y, Dangaria S, Diekwisch TG (2006) Dental follicle progenitor cell heterogeneity in the developing mouse periodontium. Stem Cells Dev 15:595–608. https://doi.org/10.1089/scd.2006.15.595
Morsczeck C et al (2005) Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol 24:155–165. https://doi.org/10.1016/j.matbio.2004.12.004
Nemoto E et al (2016) Wnt3a signaling induces murine dental follicle cells to differentiate into cementoblastic/osteoblastic cells via an osterix-dependent pathway. J Periodontal Res 51:164–174. https://doi.org/10.1111/jre.12294
Oh HJ, Lee HK, Park SJ, Cho YS, Bae HS, Cho MI, Park JC (2012) Zinc balance is critical for NFI-C mediated regulation of odontoblast differentiation. J Cell Biochem 113:877–887. https://doi.org/10.1002/jcb.23421
Pan K, Sun Q, Zhang J, Ge S, Li S, Zhao Y, Yang P (2010) Multilineage differentiation of dental follicle cells and the roles of Runx2 over-expression in enhancing osteoblast/cementoblast-related gene expression in dental follicle cells. Cell Prolif 43:219–228. https://doi.org/10.1111/j.1365-2184.2010.00670.x
Park JC, Herr Y, Kim HJ, Gronostajski RM, Cho MI (2007) Nfic gene disruption inhibits differentiation of odontoblasts responsible for root formation and results in formation of short and abnormal roots in mice. J Periodontol 78:1795–1802. https://doi.org/10.1902/jop.2007.060363
Park BW et al (2012) In vitro and in vivo osteogenesis of human mesenchymal stem cells derived from skin, bone marrow and dental follicle tissues. Differentiation 83:249–259. https://doi.org/10.1016/j.diff.2012.02.008
Park BW et al (2017) Cryopreservation of human dental follicle tissue for use as a resource of autologous mesenchymal stem cells. J Tissue Eng Regen Med 11:489–500. https://doi.org/10.1002/term.1945
Plasari G, Edelmann S, Hogger F, Dusserre Y, Mermod N, Calabrese A (2010) Nuclear factor I-C regulates TGF-β-dependent hair follicle cycling. J Biol Chem 285:34115–34125. https://doi.org/10.1074/jbc.M110.120659
Prins HJ et al (2014) In vitro induction of alkaline phosphatase levels predicts in vivo bone forming capacity of human bone marrow stromal cells. Stem Cell Res 12:428–440. https://doi.org/10.1016/j.scr.2013.12.001
Rupp RA, Kruse U, Multhaup G, Gobel U, Beyreuther K, Sippel AE (1990) Chicken NFI/TGGCA proteins are encoded by at least three independent genes: NFI-A, NFI-B and NFI-C with homologues in mammalian genomes. Nucleic Acids Res 18:2607–2616. https://doi.org/10.1093/nar/18.9.2607
Shuai Y et al (2018) Dental stem cells and tooth regeneration. Adv Exp Med Biol 1107:41–52. https://doi.org/10.1007/5584_2018_252
Sowmya S, Chennazhi KP, Arzate H, Jayachandran P, Nair SV, Jayakumar R (2015) Periodontal specific differentiation of dental follicle stem cells into osteoblast, fibroblast, and cementoblast. Tissue Eng Part C Methods 21:1044–1058. https://doi.org/10.1089/ten.TEC.2014.0603
Tan Z et al (2009) Research on promoting periodontal regeneration with human basic fibroblast growth factor-modified bone marrow mesenchymal stromal cell gene therapy. Cytotherapy 11:317–325. https://doi.org/10.1080/14653240902824757
Wise GE, Marks SJ, Cahill DR (1985) Ultrastructural features of the dental follicle associated with formation of the tooth eruption pathway in the dog. J Oral Pathol 14:15–26. https://doi.org/10.1111/j.1600-0714.1985.tb00461.x
Xiang L et al (2014) Wnt5a regulates dental follicle stem/progenitor cells of the periodontium. Stem Cell Res Ther 5:135. https://doi.org/10.1186/scrt525
Yao S, Pan F, Prpic V, Wise GE (2008) Differentiation of stem cells in the dental follicle. J Dent Res 87:767–771. https://doi.org/10.1177/154405910808700801
Yokoi T et al (2007) Establishment of immortalized dental follicle cells for generating periodontal ligament in vivo. Cell Tissue Res 327:301–311. https://doi.org/10.1007/s00441-006-0257-6
Yu X, Liu S, Chen H, Zhao X, Chen X, Du Y, Li S (2018) CGRP gene-modified rBMSCs show better osteogenic differentiation capacity in vitro. J Mol Histol 49:357–367. https://doi.org/10.1007/s10735-018-9775-2
Zhang J et al (2015) Nuclear factor I-C promotes proliferation and differentiation of apical papilla-derived human stem cells in vitro. Exp Cell Res 332:259–266. https://doi.org/10.1016/j.yexcr.2015.01.020
Zhou J et al (2017) Nuclear factor I-C reciprocally regulates adipocyte and osteoblast differentiation via control of canonical Wnt signaling. FASEB J 31:1939–1952. https://doi.org/10.1096/fj.201600975RR
Acknowledgements
This work was supported by Guangdong Science and Technology Foundation of China (Grant Number: 2014A020212086).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Zhang, F., Liang, M., Zhao, C. et al. NFIC promotes the vitality and osteogenic differentiation of rat dental follicle cells. J Mol Hist 50, 471–482 (2019). https://doi.org/10.1007/s10735-019-09841-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10735-019-09841-z