Abstract
The Arnaucho chili pepper (ACP) is a traditional vegetable used in Peru because of its gastronomic properties. Due to its importance in the Peruvian diet and economy, this species is a resource that can be a candidate to plant breeding programs. In this study, the complete genome nucleotide sequence of this chili pepper was generated using the Illumina Hiseq 2500 sequencing technology. We sequenced the whole genome of the ACP using a paired-end 150 strategy, obtaining 330.46 GB of sequencing data. The genome size of the ACP was 2.98 Gb with a contig N50 of 237 Mb and 95.39% complete BUSCOs. Also, we identified 71.96% of repetitive DNA of the genome assembly, of which retroelements occupy 37.95% of the total genome. We downloaded genomes of the Solanoideae subfamily and conducted a comparative analysis of simple sequence repeats (SSRs) with our draft genome, and we identified lower number of SSRs in the ACP genome compared to other pepper species. This first ACP genome is expected to contribute to a better understanding of its genetics to adapt to the arid conditions of the Peruvian coastal ecosystem and evolution.
Similar content being viewed by others
Introduction
The Capsicum genus species, belonging to the Solanaceae family, includes both sweet (peppers) and hot (chilies) cultivars (D’Agostino et al. 2018). It is considered a model crop due to its significant diversity, which leads to abundant phenotypic variation (Park et al. 2016). Capsicum species, initially cultivated extensively in the Americas, gained global distribution following Columbus’ voyages (Perry et al. 2007). Starch fossils of Capscium spp. were found at seven sites from the Bahamas to southern of Peru, dating back 6000 years before the first contact with Europeans (Perry et al. 2007). The USDA Genetic Resources Information Network (GRIN) currently recognized 38 Capsicum species within this genus, including five domesticated species. Some species, such as Capsicum chinense Jacq., Capsicum baccatum L. and Capsicum pubescens R. & P., are widely cultivated in South America but are less common elsewhere (Tripodi et al. 2021). Genetic, archaeological, and contemporary plant distribution evidence suggests distinct domestication origins: (i) C. chinense in the northern Amazon lowlands, (ii) C. baccatum in Bolivian lowlands, (iii) Capsicum. annuum in Mexico or northern Central America, (iv) Capsicum frutescens in the Caribbean, and (v) C. pubescens in mid-elevation Southern Andes (Perry et al. 2007; Kraft et al. 2014). These five domesticated species are facultative inbreeders, diploid, self-pollinating crops. They are closely related to tomato, potato, eggplant, petunia and tobacco (Kim et al. 2014). Capsicum species feature flowers with stellate corollas, distinct pigmentation patterns, and fleshy berries varying in size, shape, and color. A unique characteristic within the Solanaceae family is their entire cup shaped calyx (Carrizo García et al. 2016).
The Capsicum genus has been an important dietary component throughout the New World (Russo 2012) and is widely used as a natural food colorant, as an ingredient in pharmaceuticals, and as a very spicy extract used for self-defense sprays, animal repellents, and insecticides (Bosland 2016). Also, hot chili pepper provides many essential nutrients, vitamins, minerals that have great importance for human health. The most important character in Capsicum is the pungency of the fruit. This is due to the production of capsaicinoids, a group of alkaloids that are synthesized in the placenta of the fruits (Stewart et al. 2007). Capsaicinoids possess health benefits for humans and are considered anticancer (Mori et al. 2006; Ito et al. 2004), analgesic for arthritis and other pain (Fraenkel et al. 2004), and reduce appetite (Westerterp-Plantenga et al. 2005).
Next-generation sequencing (NGS) technologies speed the generation of high-throughput data (Park et al. 2016; Kim et al. 2014; Jo et al. 2011). With this technology several investigations have been conducted. (Jo et al. 2011) an analyzed the complete chloroplast genome sequence of C. annuum. Their results identified large indels, including insertions in accD and rpl20 gene sequences, and they also dentified frequent repeated tandem sequences. Kim et al (2014) reported the whole-genome sequencing and assembly of a Mexican landrace of C. annuum. They also resequenced two cultivated peppers and de novo sequenced species C. chinense and found that the genome size of the hot chili pepper was about four times larger than that of its close relative, the tomato. They also identified that the genome showed an accumulation of elements of the Gypsy and Caulimoviridae families.
The genome assembly process plays a fundamental role in genomics, where computational tools called assemblers are used to reconstruct genomic sequences from sequencing data. These assemblers vary in their computational approaches and strategies, which can significantly influence their performance across different types of data and environmental conditions (Forouzan et al. 2018). By understanding the strengths and limitations of each assembler, researchers can make informed decisions on selecting tools for their genomic analysis projects (Forouzan et al. 2017).
Unfortunately, genomic research employing the Peruvian Capsicum germplasm is still very limited. Here we report the first genome sequence of the arnaucho chili pepper, a landrace cultivated in northern Lima. This autochthonous cultivar is widely used in the Peruvian gastronomy and represents cultural and economic importance. Deciphering the ACP genome provides an evolutionary view of the expansion of the genome, also it will be possible to develop molecular markers that allow the selection of superior individuals in early stages. This would be the first step for establishing modern genetic breeding programs in Peru. On the other hand, we will know the location of the underlying genes or chromosomes that could be useful for introgression of traits of interest. The analysis of the ACP genome will be applied in the future to implement the gene editing methodology, which has resulted in impressive results in the world in favor of the development of new cultivars to improve the horticultural, nutritional and medicinal values of Capsicum species.
Materials and methods
Sample collection and DNA extraction
We collected leaves of Arnaucho chili pepper from Supe Valley, northern of Lima department (− 10.8099889, − 77.6953950). Young fresh leaves were stored in paper envelopes in plastic containers with silicone gel, for the preservation of samples during transport to the National Institute of Agricultural Innovation (INIA for its acronym in Spanish) for genomic DNA extraction. This specimen was deposited at the Germplasm Bank of INIA (https://www.gob.pe/inia, drgb@inia.gob.pe) under the voucher number PER1002642. Genomic DNA was extracted using the CTAB method (Doyle and Doyle 1987) with slightly modifications for this species. DNA quantity was evaluated using the Qubit™4 Fluorometer (Invitrogen,Waltham, MA, USA) and quality was evaluated in agarose gel (1%).
DNA sequencing and estimation of genome size
Pair-end DNA sequencing was conducted using the Illumina HiSeq 2500 system. Read quality was assessed by FastQC v.0.11.9 software (Andrews 2014). Also, the reads were trimmed and filtered (phred Q > 25) using Trimmomatic v0.36 (Bolger et al. 2014) and TrimGalore v.0.6.10 software (Krueger 2012) with default parameters. We employed the Illumina short-read data and the k-mer analysis with programs Jellyfish v.2.0. and Genome Scope v1.0.0 (Cold Spring Harbor Laboratory, Laurel Hollow, US) (Vurture et al. 2017) to estimate the features of the genome, including genome size, repeat content, and heterozygosity rate. K-depth was estimated to identify a common single-peak pattern in the k-mer frequency distribution analysis.
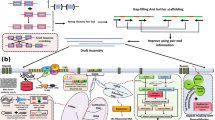
Genome assembly
De novo assembly was performed with two assembly algorithms: SOAPdenovo2 v.2.04 (Luo et al. 2012), and MaSuRCA v.4.0.6 (Zimin et al. 2013). These assemblers were selected due to their distinct methodological approaches, enabling a comparative evaluation to identify the most suitable algorithm for this genome. SOAPdenovo2 rapidly assembles short reads into contigs with high efficiency, while MaSuRCA employs a hybrid approach to generate longer contigs and scaffolds, particularly in complex genomic regions.. Next, we used QUAST v.5.2.0 for statistics of assemblies. MaSuRCA resulted in improved assembly statistics and was subjected to Samba Scaffolder v.1.0 [21] for scaffolding and gap-filling. For the reference-based scaffolding, we used C. chinense genome (Genbank: GCA_002271895.2). Next, we used QUAST with the output of the scaffolding. Validation of assembly was assessed using two different approaches: (i) filtered PE Illumina reads were remapped to detect errors in the assembly using Bowtie2 v.2.4.2 (Langmead and Salzberg 2012) and SamTools v.1.7 (Li et al. 2009) software, (ii) the BUSCO (Simão et al. 2015) strategy was used to test the completeness of the genome assembly and gene space using the mammalian-specific profile. This approach makes use of single-copy genes expected to be present in plants (4,104 genes). We used JCVI vecscreen (https://github.com/tanghaibao/jcvi), which uses Univecdatabase (https://ftp.ncbi.nlm.nih.gov/pub/UniVec/) for detection of vectors, and 900 mapped the scaffolds against to nt/nr National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/) using BLAST v.2.2.26 (Altschult et al. 1990) for identifying contamination. The mitochondrial DNA sequences were removed after BLAST searches against databases of mitochondrial sequences. Finally, we discarded contaminated scaffolds and vectors to submit to the NCBI database.
Repeat annotation and SSR Data mining
To identify repetitive elements, we used de novo and homolog-based methods. For the de novo approach, we used, with default parameters, RepeatModeler v.1.08 (Abrusán et al. 2009) to generate a de novo ACP repeat library, which is subsequently used in RepeatMasker v4.0.7 (Bedell et al. 2000) to annotate repeats. For the homology-based approaches, we used Repbase v4.0.7 (Bao et al. 2015), RepeatMasker and RMBlast v2.2.27 (Korf 2004). All repeat results were merged, and final summary was reported using RepeatMasker. The SSRs were identified in the genome using MISA perl script (http://pgrc.ipk-gatersleben.de/misa/) (Beier et al. 2017) with the specific settings: monomer (one nucleotide, n > 12), dimer (two nucleotides, n > 6), trimer (three nucleotides, n > 4), tetramer (four nucleotides, n > 3), pentamer (five nucleotides, n > 3), and hexamer (six nucleotides, n > 3). Also, for SSR comparison, we added the genomes from Capsicum rhomboideum (GenBank:GCA_031232105.1), C. annuum (GenBank: GCA_002878395.3), C. baccatum var. pendulum (GenBank: GCA_030864225.1), C. pubescens (GenBank: GCA_030864215.1), C. chinense (Genbank: GCA_026120095.1), C. annuum (Genbank: GCA_021292125.1), C. annuum var. annuum (Genbank: GCA_033026575.1), C. annuum var. glabriusculum (Genbank: GCA_000950795.1), and C. baccatum (Genbank: GCA_002271885.2). Afterward, we used the MISA script with the same parameters for ACP.
Results
The whole genome sequencing data was deposited in the Short Read Archive (SRA) database under accession number SRR21189431, Biosample SAMN30481898, Bioproject PRJNA873099. The total of raw pair-reads were 1,062,221,314 sequences with mean length of 150 pb, 35% GC content, and a total output of 330.467 GB sequencing data. After the trimming, we retained 95% of sequencing data; 1,008 million high-quality sequence reads with approximate 313 GB total sequencing data were generated. We did not detect overrepresented sequences and trimmed adapters. Also, the average quality per read was 36.
Genomic survey
We obtained a low heterozygosity (0.078429%), slightly repetitive (37.6%), and the estimation of the genome (2.358 Gb) was relatively close to the reported references of genomes for C. chinense (Genbank:GCA_002271895.2; 3.07 Gb) (Table 1). Based on the estimated draft genome size, subsequent de novo assembly and genome annotation were performed with the sequencing depth of approximately 63.06 X coverage (Fig. 1).
Assembly de novo and validation
We obtained different assemblies from SOAPdenovo2 and MaSuRCA programs. We continued our analysis with MaSuRCA due to a better N50 and longer contigs (Supplementary Data 1). Assembly MaSuRCA genome was scaffolded, and we obtained a total length of 2.98 Gb, which had 11,930 contigs (≥ 1000 bp) with a GC content of 34.99%. The longest contig was of 275,254,545 pb. In addition, we found that 99.33% of the raw paired-end reads generated from the small insertion libraries were aligned to our final assembled genome. Also, with the scaffolding approach, we improved the N50, from 13.22 kb to 237.34 Mb (Table 1). In the Solanales order, the number of complete single-copy BUSCOs (Benchmarking Universal Single-Copy) increased from 5201 to 5895 complete BUSCOs (Table 2), and the number of fragmented BUSCOs decreased from 951 to 139. The genome sequence is openly available in the Genbank of NCBI under the accession number JAPEIP000000000.1.
Simple sequence repeat data mining
The predominant microsatellite motif in the Arnaucho genome comprised mononucleotide repeats, constituting 57.74% of the total SSRs. Dinucleotide repeats accounted for 28.52%, trinucleotide repeats for 14.69%, and tetranucleotide repeats for 1.80%. This distribution closely mirrors the microsatellite motif patterns observed in other pepper species, including GCA_000710875.1, GCA_002271885.2, GCA_000950795.1, GCA_033026575.1, GCA_030864215.1, GCA_030864225.1, GCA_002878395.3, and GCA_031323105.1 (Fig. 2).
A total of 590,103 microsatellite loci were identified in the assembled Arnaucho draft genome sequence, showing a frequency of 0.14 SSR/Mb. This frequency is comparable to GCA_000950795.1 (0.18 SSR/Mb) but lower than GCA_000710875.1 (0.2 SSR/Mb), GCA_002271885.2 (0.19 SSR/Mb), GCA_033026575.1 (0.22 SSR/Mb), GCA_030864215.1(0.22 SSR/Mb), GCA_030864225.1 (0.23 SSR/Mb), GCA_002878395.3 (0.2 SSR/Mb), and GCA_031323105.1 (0.23 SSR/Mb) (Table 3). Additionally, the composite matrix of the Arnaucho genome contains a total of 76,301 SSRs, which is lower than all others.
Discussion
The selection of sequencing technique and assembly program played a crucial role in generating and ensuring the quality of the assembled genome of Arnaucho chili pepper. The importance of assembly program selection in the quality of the assembled genome has been previously highlighted (Forouzan et al. 2018), and significant differences in the quality and extent of the assembled genome were observed in this study through the contrast between the SOAPdenovo2 and MaSuRCA assembly methods. The internal results of this study demonstrated that MaSuRCA produced an improved N50 and longer contigs compared to SOAPdenovo2, leading to the continuation of the analysis with MaSuRCA. This observation underscores the significant influence of assembly program selection on the quality and extent of the assembled genome (Martínez-García et al. 2016), emphasizing the importance of this stage in obtaining a representative and reliable genome for subsequent analysis and annotation.
The genomes of Capsicum annuum and C. chinense exhibited a notably high proportion of divergence, indicating considerable genetic variation between these species. Additionally, the Arnaucho chili pepper genome showed a higher abundance of transposable elements (TEs), including long terminal repeat (LTR) elements and elements from the Caulimoviridae family, compared to the pepper genome. These TEs are widely dispersed throughout the Arnaucho chili pepper genome and significantly contribute to its expansion. Furthermore, there is a differentiation in the accumulation of Gypsy and Copia elements between both species, potentially contributing to the speciation of the Arnaucho chili pepper (Kim et al. 2014).
The comprehensive analysis of repetitive DNA in the Arnaucho chili pepper genome reveals a complex and diverse dynamics of retrotransposable elements within its genome. Retroelements represent a significant fraction of repetitive sequences, with LTRs being the major fraction, occupying between 20% and 80% of the genome depending on the species (Yañez-Santos et al. 2021). The prevalence of TEs, notably LTR retrotransposons, underscores the substantial impact of these elements on the repetitive sequences. Furthermore, the observed variability in the proportion of LTRs among species underscores the genomic diversity within Capsicum (De Assis et al. 2020). This variation in retroelement quantity has been identified as one of the main contributors to the variability in the genomic size of the botanical family Solanaceae (Park et al. 2011). The presence of complete and intact retroelements, constituting a minor fraction of the genome, reveals their ability to induce changes in genomic structure and promote genome divergence and evolution (Yañez-Santos et al. 2021). Notably, the Gypsy retrotransposon emerges as one of the most abundant elements in the repetitive DNA of the Arnaucho chili pepper, reinforcing its significance in the genomic configuration and structure of this species. These findings align with comparative research between chili pepper and tomato, underscoring the importance of differential accumulation of repetitive elements in the expansion of euchromatic regions (De Assis et al. 2020).
The uniformity in the distribution of microsatellite motifs shared with other pepper cultivars such as Pepper Zunla 1, Pepper Chiltepin, and those mentioned previously, suggests coherence in the genomic patterns of microsatellites within the Capsicum genus. This could indicate a conservation of these genetic patterns, revealing a crucial aspect for genomic stability (Li et al. 2002) within the Capsicum genus.
The microsatellite frequency in this genome, expressed as 0.14 SSR/Mb, provides an enlightening measure of the density of these sequences in its genome. This variable becomes even more significant when contrasted with other cultivars, highlighting variability in the abundance of microsatellites among the studied species, suggesting potential differences in genomic structure and genetic diversity (Srivastava et al. 2019; Fischer et al. 2017) among pepper species. Regarding the lower number of SSRs in the Arnaucho genome compared to other pepper cultivars, several factors can be considered. It is possible that evolutionary processes specific to the Arnaucho cultivar have led to a reduction in the number of SSRs, potentially through selection pressures or genetic drift (Bagshaw 2017). Further research into the genomic and evolutionary mechanisms underlying these differences could provide valuable information on the genetic diversity and adaptation of pepper cultivars.
Conclusion
The selection of sequencing techniques and assembly programs played a pivotal role in shaping the quality and comprehensiveness of the assembled Arnaucho chili pepper genome. The preference for MaSuRCA assembler was based on superior metrics like N50 and contig length, highlighting the significance of program choice in obtaining a reliable genome for subsequent analyses. Additionally, the consistent distribution of microsatellite motifs across diverse Capsicum species, including Arnaucho, hints stable genomic patterns within this genus, potentially preserving essential genetic elements for genomic stability. Furthermore, the observed variability in microsatellite abundance, highlighted by microsatellite frequency of 0.14 SSR/Mb compared to other studied species, implies potential distinctions in genomic structure and genetic diversity among pepper species. These findings offer crucial insights into the genomic intricacies of Capsicum species, providing a foundation for further explorations and understanding within this field.
Data availability statement
All data generated during this study are included in this published article.
References
Abrusán G, Grundmann N, Demester L, Makalowski W (2009) TEclass—a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 25:1329–1330. https://doi.org/10.1093/bioinformatics/btp084
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Andrews S (2014) FastQC a quuality control tool for high throughput sequence data. Bioinformatics 30:2114–2120
Bagshaw ATM (2017) Functional Mechanisms of Microsatellite DNA in Eukaryotic Genomes. Genome Biol Evol 9:2428–2443. https://doi.org/10.1093/gbe/evx164
Bao W, Kojima KK, Kohany O (2015) Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. https://doi.org/10.1186/s13100-015-0041-9
Bedell JA, Korf I, Gish W (2000) MaskerAid: a performance enhancement to RepeatMasker. Bioinformatics 16:1040–1041. https://doi.org/10.1093/bioinformatics/16.11.1040
Beier S, Thiel T, Münch T et al (2017) MISA-web: a web server for microsatellite prediction. Bioinformatics 33:2583–2585. https://doi.org/10.1093/bioinformatics/btx198
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bosland PW (2016) Capsicums: innovative uses of an ancient crop. In: Janick, J., Ed., Progress in New Crops ASHS Press, Arlington. Agric Trop Subtrop 479–487. https://doi.org/10.4236/oalib.1102396
Carrizo García C, Barfuss MHJ, Sehr EM et al (2016) Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann Bot 118:35–51. https://doi.org/10.1093/aob/mcw079
D’Agostino N, Tamburino R, Cantarella C et al (2018) The complete plastome sequences of eleven capsicum genotypes: insights into DNA variation and molecular evolution. Genes (basel). https://doi.org/10.3390/genes9100503
De Assis R, Baba VY, Cintra LA et al (2020) Genome relationships and ltr-retrotransposon diversity in three cultivated capsicum 1. (solanaceae) species. BMC Genom. https://doi.org/10.1186/s12864-020-6618-9
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Fischer MC, Rellstab C, Leuzinger M et al (2017) Estimating genomic diversity and population differentiation—an empirical comparison of microsatellite and SNP variation in Arabidopsis halleri. BMC Genom. https://doi.org/10.1186/s12864-016-3459-7
Forouzan E, Maleki MSM, Karkhane AA, Yakhchali B (2017) Evaluation of nine popular de novo assemblers in microbial genome assembly. J Microbiol Methods 143:32–37. https://doi.org/10.1016/j.mimet.2017.09.008
Forouzan E, Shariati P, Mousavi Maleki MS et al (2018) Practical evaluation of 11 de novo assemblers in metagenome assembly. J Microbiol Methods 151:99–105. https://doi.org/10.1016/j.mimet.2018.06.007
Fraenkel L, Bogardus ST, Concato J, Wittink DR (2004) Treatment options in knee osteoarthritis the patient’s perspective. Arch Intern Med 164:1299–1304
Ito K, Nakazato T, Yamato K et al (2004) (2004) Induction of apoptosis in leukemic cells by homovanillic acid derivative, capsaicin, through oxidative stress: implication of phosphorylation of p53 at ser-15 residue by reactive oxygen species. Cancer Res 64(3):1071–1078
Jo YD, Park J, Kim J et al (2011) Complete sequencing and comparative analyses of the pepper (Capsicum annuum L.) plastome revealed high frequency of tandem repeats and large insertion/deletions on pepper plastome. Plant Cell Rep 30:217–229. https://doi.org/10.1007/s00299-010-0929-2
Kim S, Park M, Yeom SI et al (2014) Genome sequence of the hot pepper provides insights into the evolution of pungency in capsicum species. Nat Genet 46:270–278. https://doi.org/10.1038/ng.2877
Korf I (2004) Gene finding in novel genomes. BMC Bioinformatics 14:5–59
Kraft KH, Brown CH, Nabhan GP, Luedeling E, Luna Ruiz JDJ, Eeckenbrugge d’ Coppens G, Gepts P (2014) Multiple lines of evidence for the origin of domesticated chili pepper, Capsicum annuum, in Mexico. Proc Natl Acad Sci USA 111:6165–6170
Krueger, F., (2012). Trim Galore. Babraham Bioinformatics. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Li YC, Korol AB, Fahima T et al (2002) Microsatellites: genomic distribution, putative functions and mutational mechanisms: a review. Mol Ecol 11:2453–2465
Li H, Handsaker B, Wysoker A et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Luo R, Liu B, Xie Y, Li Z, Huang W et al (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18
Martínez-García PJ, Crepeau MW, Puiu D et al (2016) The walnut (Juglans regia) genome sequence reveals diversity in genes coding for the biosynthesis of non-structural polyphenols. Plant J 87:507–532. https://doi.org/10.1111/tpj.13207
Mori A, Lehmann S, O’Kelly J et al (2006) Capsaicin, a component of red peppers, inhibits the growth of androgen-independent, p53 mutant prostate cancer cells. Cancer Res 66:3222–3229. https://doi.org/10.1158/0008-5472.CAN-05-0087
Park M, Jo SH, Kwon JK et al (2011) Comparative analysis of pepper and tomato reveals euchromatin expansion of pepper genome caused by differential accumulation of Ty3/Gypsy-like elements. BMC Genom. https://doi.org/10.1186/1471-2164-12-85
Park HS, Lee J, Lee SC et al (2016) The complete chloroplast genome sequence of Capsicum chinense Jacq. (Solanaceae). Mitochondrial DNA B Resour 1:164–165. https://doi.org/10.1080/23802359.2016.1144113
Perry L, Dickau R, Zarrillo S et al (1979) (2007) Starch fossils and the domestication and dispersal of chili peppers (Capsicum spp. L.) in the Americas. Science 315:986–988. https://doi.org/10.1126/science.1136914
Russo V (2012) Peppers: botany, production and uses. CAB International, London
Simão FA, Waterhouse RM, Ioannidis P et al (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. https://doi.org/10.1093/bioinformatics/btv351
Srivastava S, Avvaru AK, Sowpati DT, Mishra RK (2019) Patterns of microsatellite distribution across eukaryotic genomes. BMC Genom. https://doi.org/10.1186/s12864-019-5516-5
Stewart C, Mazourek M, Stellari GM et al (2007) Genetic control of pungency in C. chinense via the Pun1 locus. J Exp Bot 58:979–991. https://doi.org/10.1093/jxb/erl243
Tripodi P, Rabanus-Wallace MT, Barchi L, Kale S et al (2021) Global range expansion history of pepper (Capsicum spp.) revealed by over 10,000 genebank accessions. Proc Natl Acad Sci 118:2104315118. https://doi.org/10.1073/pnas.2104315118
Vurture GW, Sedlazeck FJ, Nattestad M et al (2017) GenomeScope Fast reference-free genome profiling from short reads. Bioinformatics. Oxford University Press, Oxford, pp 2202–2204
Westerterp-Plantenga MS, Smeets A, Lejeune MPG (2005) Sensory and gastrointestinal satiety effects of capsaicin on food intake. Int J Obes 29:682–688. https://doi.org/10.1038/sj.ijo.0802862
Yañez-Santos AM, Paz RC, Paz-Sepúlveda PB, Urdampilleta JD (2021) Full-length LTR retroelements in Capsicum annuum revealed a few species-specific family bursts with insertional preferences. Chromosome Res 29:261–284. https://doi.org/10.1007/s10577-021-09663-4
Zimin AV, Marçais G et al (2013) The MaSuRCA genome assembler. Bioinformatics. 29:2669–2677. https://doi.org/10.1093/bioinformatics/btt476
Acknowledgements
We thank Ivan Ucharima for supporting the logistic activities in the laboratory. Finally, the authors thank the Bioinformatics High-Performance Computing server of La Molina National Agrarian University (UNALM) for providing computational resources for data analysis. C.I.A. thanks Vicerrectorado de Investigación of UNTRM.
Funding
This research was funded by PP068 “Reducción de la vulnerabilidad y atención de emergencias por desastres” of the Ministry of Agrarian Development and Irrigation (MIDAGRI) of the Peruvian Government.
Author information
Authors and Affiliations
Contributions
Conceptualization, RE, JFC and CIA; methodology, RE and CS; software, RE; validation, RE, YR; formal analysis, RE and JFC; investigation, RE; resources, CS and CIA; data curation, RE, and JFC; writing—original draft preparation, RE, EV, and YR; writing—review and editing, RE, JFC, CS and CIA; visualization, RE, and JFC; supervision, CS, and CIA; project administration, CS, and CIA; funding acquisition, CIA. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Estrada, R., Tantalean, J.F.C., Saldaña, C.L. et al. Draft genome and SSR data mining of a Peruvian landrace of Capsicum chinense, the arnaucho chili pepper. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-01941-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-01941-4