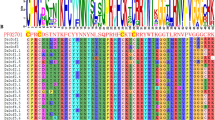
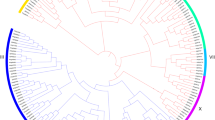
Abstract
DOF transcription factors are essential plant-specific gene regulators that are crucial in plant growth and stress responses. Mangifera indica, a tropical fruit species with diverse applications, undergoes complex genetic regulation during fruit ripening, influenced by climacteric conditions. This study focuses on DNA-binding one zinc finger genes (MiDofs) in Mangifera indica (mango) and their response to drought stress during fruit development. We found 36 MiDof genes in mango, categorizing them into six distinct clusters based on evolutionary relationships. Despite variations in various characteristics, the fundamental structures of these MiDof transcription factors remained consistent. Notably, two MiDof genes were outside the nucleus. Gene structure analysis and motif identification revealed common features among all MiDofs, with certain genes like MiDof5.3 and MiDof2.2 performing unique functions. Our analysis of conserved domains highlighted crucial functional sites and protein–protein interaction networks, with the zf-Dof domain being the most conserved. Additionally, we explored cis-acting regulatory elements, shedding light on the mechanisms by which MiDofs respond to light and drought stress. Expression analysis demonstrated the involvement of MiDofs in mango fruit ripening and development under light conditions. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) confirmed the induction of MiDofs under drought stress when compared to a control group. This study contributes valuable insights to the field of fruit biology and provides a deeper understanding of the role of MiDof genes in mango fruit development, offering potential avenues for crop improvement strategies.






Similar content being viewed by others
Data availability
All data generated or analyzed analyzed during this study are included in this published article.
Abbreviations
- Dof :
-
DNA-binding One Zinc Finger
- GSDS:
-
Gene Structure Display Server
- GRAVY:
-
Average hydropathy
- pI:
-
Isoelectric point
- AI:
-
Aliphatic index
- PPI:
-
Protein–protein interactions
- FPKM:
-
Fragments per kilobase of transcript per million mapped reads
References
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43:39–49
Basu S, Ramegowda V, Kumar A, Pereira A (2016) Plant adaptation to drought stress. F1000Research 5:1554. https://doi.org/10.12688/f1000research.7678.1
Biswas B, Kumar L (2011) Revolution of Mango production. Fertil Mark News 1:1–24
Bustin SA, Mueller R (2005) Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin Sci 109:365–379
Chadha K, Pal R (2018) Mangifera indica. CRC Press, CRC handbook of flowering
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, HeYXia R (2020) TB tools: an integrative toolkit developed for interactive analyses of big biological data. Mol plant, 13: 1194–1202 Consortium MG, Bally IS, Bombarely A, Chambers AH, Cohen Y, Dillon NL, Innes DJ, Islas-Osuna M A, Kuhn DN, Mueller LA (2021) The ‘Tommy Atkins’ mango genome reveals candidate genes for fruit quality. BMC Plant Biol 21:1–18
Das MK, Dai H-K (2007) A survey of DNA motif finding algorithms. BMC Bioinformatics 8:1–13
Fang Z, Jiang W, He Y, Ma D, Liu Y, Wang S, Zhang Y, Yin J (2020) Genome-wide identification, structure characterization, and expression profiling of Dof transcription factor gene family in wheat (Triticum aestivum L.). Agronomy 10:294
Farooq M, Hussain M, Wahid A, Siddique K (2012) Drought Stress in Plants: An Overview. In: Aroca, R (eds) Plant Responses to Drought Stress. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-32653-0_1
Guo Y, Qiu L-J (2013) Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 8:e76809
Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297
Jahurul M, Zaidul I, Ghafoor K, Al-Juhaimi FY, Nyam KL, Norulaini N, SahenaOmar FAM (2015) Mango (Mangifera indica L.) by-products and their valuable components: a review. Food Chem 183:173–180
Khaksar G, Sangchay W, Pinsorn P, SangpongSirikantaramas LS (2019) Genome-wide analysis of the Dof gene family in durian reveals fruit ripening-associated and cultivar-dependent Dof transcription factors. Scient Rep 9:12109
Kibbe WA (2007) OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res 35:W43–W46
Kushwaha H, Gupta S, Singh VK, Rastogi S, Yadav D (2011) Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and arabidopsis. Mol Biol Rep 38:5037–5053
Lehti-Shiu MD, Panchy N, Wang P, Uygun S, Shiu SH (2017) Diversity, expansion, and evolutionary novelty of plant DNA-binding transcription factor families. Biochim Biophys Acta Gene Regul Mech 1860:3–20
Leinonen R, Sugawara H, Shumway M, Collaboration INSD (2010) The sequence read archive. Nucleic Acids Res 39:D19–D21
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van De Peer Y, Rouzé P, Rombauts S (2002) Plant Care, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Letunic I, Bork P (2019) Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259
Lijavetzky D, Carbonero P, Vicente-Carbajosa J (2003) Genome-wide comparative phylogenetic analysis of the rice and arabidopsis Dof gene families. BMC Evol Biol 3:1–11
Lohani N, Babaei S, SinghBhalla MBPL (2021) Genome-wide in silico identification and comparative analysis of Dof gene family in Brassica napus. Plants 10:709
Mahram A, Herbordt MC (2010) Fast and accurate NCBI BLASTP: acceleration with multiphase FPGA-based prefiltering. In: Proceedings of the 24th ACM International conference on supercomputing 73–82
Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI (2015) CDD: NCBI’s conserved domain database. Nucleic Acids Res 43:D222–D226
Mering CV, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B (2003) STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 31:258–261
Nadeem SM, Ahmad M, Zahir ZA, Javaid A, Ashraf M (2014) The role of mycorrhizae and plant growth promoting rhizobacteria (PGPR) in improving crop productivity under stressful environments. Biotechnol Adv 32:429–448
Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ (2015) Iq-tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274
Noratto GD, Bertoldi MC, Krenek K, Talcott ST, Stringheta PC, Mertens-Talcott SU (2010) Anticarcinogenic effects of polyphenolics from mango (Mangifera indica) varieties. J Agric Food Chem 58:4104–4112
Parvez GM (2016) Pharmacological activities of mango (Mangifera indica): a review. J Pharm Phytoch 5:01–07
Poole RL (2007) The TAIR database methods and protocols. Plant Bioinfo 1:179–212
Rowe N, Speck T (2005) Plant growth forms: an ecological and evolutionary perspective. New Phytol 166:61–72
Shah K, Patel M, Patel R, Parmar P (2010) Mangifera indica (mango). Pharmacogn Rev 4:42
Shu Y, Song L, Zhang J, Liu Y, Guo C (2015) Genome-wide identification and characterization of the Dof gene family in medicago truncatula. Gen Mol Res 14:10645–10657
Singh L (1960) The mango botany. Cultivation and Utilization Inter-Science Publication Inc, New York
Song A, Gao T, Li P, Chen S, Guan Z, Wu D, Xin J, Fan Q, Zhao K, Chen F (2016) Transcriptome-wide identification and expression profiling of the DOF transcription factor gene family in Chrysanthemum morifolium. Front Plant Sci 7:199
Tharanathan R, YashodaPrabha HT (2006) Mango (Mangifera indica L.), “The king of fruits”—An overview. Food Rev Int 22:95–123
Wang Wang Y, Tong Q, Xu G, Xu M, Li H, Fan P, Li S, Liang Z (2021) Transcriptomic analysis of grapevine Dof transcription factor gene family in response to cold stress and functional analyses of the VaDof17d gene. Planta 253:1–14
Yanagisawa S (2002) The Dof family of plant transcription factors. Trends Plant Sci 7:555–560
Yang X, Tuskan GA, Cheng ZM (2006) Divergence of the Dof gene families in poplar, arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol 142:820–830
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, RozenMadden RTL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. Bioinformatics 13:1–11
Zou Z, Zhu J, Zhang X (2019) Genome-wide identification and characterization of the Dof gene family in cassava (Manihot esculenta). Gene 687:298–307
Acknowledgements
Princess Nourah bint Abdulrahman University researchers Supporting Project number (PNURSP2023R294), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding
Princess Nourah bint Abdulrahman University researchers Supporting Project number (PNURSP2023R294), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
All the authors contributed to the study conception and design. SMSA, AA, MAA, MHS and BMA involved in conceptualization and supervision. MAA for funding acquisition. Material preparation, data collection and analysis were performed by NAA, MAA, SMSA, GSHA and DA. BMA, DA and MAA, KMA provided technical support during the analysis. The first draft of the manuscript was written by DAA, DA, SMAQ and MHS and all the authors commented on previous versions of the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Research involving human participants and/or animals
As a corresponding author, I confirm that our research don’t involve human participants and/or animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Alghanem, S.M.S., Alnusairi, G.S.H., Alkhateeb, M.A. et al. Genome-wide identification and characterization of the dof gene family in mango (Mangifera indica L.). Genet Resour Crop Evol (2023). https://doi.org/10.1007/s10722-023-01767-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-023-01767-6