Abstract
The biological meaning of low complexity regions in the proteins of Plasmodium species is a topic of discussion in evolutionary biology. There is a debate between selectionists and neutralists, who either attribute or do not attribute an effect of low-complexity regions on the fitness of these parasites, respectively. In this work, we comparatively study 22 Plasmodium species to understand whether their low complexity regions undergo a neutral or, rather, a selective and species-dependent evolution. The focus is on the connection between the codon repertoire of the genetic coding sequences and the occurrence of low complexity regions in the corresponding proteins. The first part of the work concerns the correlation between the length of plasmodial proteins and their propensity at embedding low complexity regions. Relative synonymous codon usage, entropy, and other indicators reveal that the incidence of low complexity regions and their codon bias is species-specific and subject to selective evolutionary pressure. We also observed that protein length, a relaxed selective pressure, and a broad repertoire of codons in proteins, are strongly correlated with the occurrence of low complexity regions. Overall, it seems plausible that the codon bias of low-complexity regions contributes to functional innovation and codon bias enhancement of proteins on which Plasmodium species rest as successful evolutionary parasites.


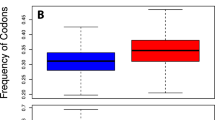
taken from GitHub-Matlab. c Trend of the sample averages of the distributions of b with respect to the average Guanine and Cytosine content of each parasite (Table 1)










Similar content being viewed by others
Data availability
Data are retrievable from public archives as mentioned in methods. Supplementary files are provided as electronically.
Code availability
Scripts are available, upon request, from the corresponding author.
References
Albà MM, Guigó R (2004) Comparative analysis of amino acid repeats in rodents and humans. Genome Res 14:549–554. https://doi.org/10.1101/gr.1925704
Arisue N et al (2019) Apicoplast phylogeny reveals the position of Plasmodium vivax basal to the Asian primate malaria parasite clade. Sci Rep 9:7274. https://doi.org/10.1038/s41598-019-43831-1
Battistuzzi FU et al (2016) Profiles of low complexity regions in Apicomplexa. BMC Evolut Biol 16:47. https://doi.org/10.1186/s12862-016-0625-0
Carter R, Walliker D (1975) New observations on the malaria parasites of rodents of the Central African Republic - Plasmodium vinckei petteri subsp. nov. and Plasmodium chabaudi Landau, 1965. Ann Trop Med Parasitol 69:187–196. https://doi.org/10.1080/00034983.1975.11687000
Castillo AI, Nelson A, Lyons E (2019) Tail wags the dog? Functional gene classes driving genome-wide gc content in plasmodium spp. Genome Biol Evol 11:497–507. https://doi.org/10.1093/gbe/evz015
Chaudhry SR et al (2018) Comparative analysis of low complexity regions in Plasmodia. Sci Rep 8:335. https://doi.org/10.1038/s41598-017-18695-y
Chen YC et al (2013) Effects of GC bias in next-generation-sequencing data on de novo genome assembly. PLoS One. https://doi.org/10.1371/journal.pone.0062856
Collins WE, Jeffery GM (2005) Plasmodium ovale: parasite and disease. Clinical microbiology reviews 18:570–581. https://doi.org/10.1128/CMR.18.3.570-581.2005
Collins WE, Jeffery GM (2007) Plasmodium malariae: parasite and disease. Clin Microbiol Rev 20:579–592. https://doi.org/10.1128/CMR.00027-07
Corradetti A, Garnham PCC, Laird M (1963) New classification of the avian malaria parasites. Parassitologia 5:1–4
Deiana A et al (2019) Intrinsically disordered proteins and structured proteins with intrinsically disordered regions have different functional roles in the cell. PLoS One. https://doi.org/10.1371/journal.pone.0217889
DePristo MA, Zilversmit MM, Hartl DL (2006) On the abundance, amino acid composition, and evolutionary dynamics of low-complexity regions in proteins. Gene 378:19–30. https://doi.org/10.1016/j.gene.2006.03.023
Duret L, Mouchiroud D (1999) Expression pattern and surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proceed Natl Acad Sci USA 96:4482–4487. https://doi.org/10.1073/pnas.96.8.4482
Escalante AA, Ayala FJ (1994) Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proceed Natl Acad Sci USA 91:11373–11377. https://doi.org/10.1073/pnas.91.24.11373
Faux NG et al (2005) Functional insights from the distribution and role of homopeptide repeat-containing proteins. Genome Res 15:537–551. https://doi.org/10.1101/gr.3096505
Ferreira et al (2003) Sequence diversity and evolution of the malaria vaccine candidate merozoite surface protein-1 (MSP-1) of Plasmodium falciparum. Gene 304:65–75. https://doi.org/10.1016/s0378-1119(02)01180-0
Filisetti et al (2013) Aminoacylation of Plasmodium falciparum tRNA (Asn) and insights in the synthesis of asparagine repeats. J Biol Chem 288:36361–36371. https://doi.org/10.1074/jbc.M113.522896
Forcelloni S, Giansanti A (2020) Evolutionary forces and codon bias in different flavors of intrinsic disorder in the human proteome. J Mol Evol 88:164–178. https://doi.org/10.1007/s00239-019-09921-4
Forsdyke D (2016) Evolutionary Bioinformatics. Springer
Frugier M et al (2010) Low complexity regions behave as tRNA sponges to help co-translational folding of plasmodial proteins. FEBS Letters 584:448–454. https://doi.org/10.1016/j.febslet.2009.11.004
Gajbhiye S, Patra PK, Yadav MK (2017) New insights into the factors affecting synonymous codon usage in human infecting Plasmodium species. Acta Trop 176:29–33. https://doi.org/10.1016/j.actatropica.2017.07.025
Garnham PC (1964) The subgenera of plasmodium in mammals. Ann Soc Belges Med Trop Parasitol Mycol 44:267–271
Gemayel R et al (2012) Beyond junk-variable tandem repeats as facilitators of rapid evolution of regulatory and coding sequences. Genes 3:461–480. https://doi.org/10.3390/genes3030461
Gragg H, Harfe BD, Jinks-Robertson S (2002) Base composition of mononucleotide runs affects DNA polymerase slippage and removal of frameshift intermediates by mismatch repair in Saccharomyces cerevisiae. Mol Cel Biol 22:8756–8762. https://doi.org/10.1128/mcb.22.24.8756-8762.2002
Hamilton WL et al (2017) Extreme mutation bias and high AT content in Plasmodium falciparum. Nucleic Acids Res 45:1889–1901. https://doi.org/10.1093/nar/gkw1259
Hou N et al (2020) Low-complexity repetitive epitopes of plasmodium falciparum are decoys for humoural immune responses. Front Immunol 11:610. https://doi.org/10.3389/fimmu.2020.00610
Howes RE et al (2016) Global epidemiology of Plasmodium vivax. Am J Trop Med Hygiene 95:15–34. https://doi.org/10.4269/ajtmh.16-0141
Karlin S et al (2002) Amino acid runs in eukaryotic proteomes and disease associations. Proceed Natl Acad Sci USA 99:333–338. https://doi.org/10.1073/pnas.012608599
Kebede AM et al (2019) Effect of low complexity regions within the PvMSP3α block II on the tertiary structure of the protein and implications to immune escape mechanisms. BMC Str Biol 19:6. https://doi.org/10.1186/s12900-019-0104-0
Kristan M et al (2019) Mosquito and human hepatocyte infections with Plasmodium ovale curtisi and Plasmodium ovale wallikeri. Trans Royal Soc Trop Med Hygiene 113:617–622. https://doi.org/10.1093/trstmh/trz048
Kristofich J et al (2018) Synonymous mutations make dramatic contributions to fitness when growth is limited by a weak-link enzyme. PLoS Genet. https://doi.org/10.1371/journal.pgen.1007615
Larson B (2019) Origin of Two Most Virulent Agents of Human Malaria: Plasmodium falciparum and Plasmodium vivax. https://doi.org/10.5772/intechopen.84481
Legendre M et al (2007) Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res 17:1787–1796. https://doi.org/10.1101/gr.6554007
Levinson G, Gutman GA (1987) Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol 4:203–221. https://doi.org/10.1093/oxfordjournals.molbev.a040442
Muralidharan V, Goldberg DE (2013) Asparagine repeats in Plasmodium falciparum proteins: good for nothing? PLoS Pathogens. https://doi.org/10.1371/journal.ppat.1003488
Muralidharan V et al (2012) Plasmodium falciparum heat shock protein 110 stabilizes the asparagine repeat-rich parasite proteome during malarial fevers. Nat Commun 3:1310. https://doi.org/10.1038/ncomms2306
Novembre JA (2002) Accounting for background nucleotide composition when measuring codon usage bias. Mol Biol Evol 19:1390–1394. https://doi.org/10.1093/oxfordjournals.molbev.a004201
Otto TD et al (2018) Genomes of all known members of a Plasmodium subgenus reveal paths to virulent human malaria. Nat Microbiol 3:687–697. https://doi.org/10.1038/s41564-018-0162-2
Pizzi E, Frontali C (2001) Low-complexity regions in Plasmodium falciparum proteins. Genome Res 11:218–229. https://doi.org/10.1101/gr.gr-1522r
Plotkin JB, Kudla G (2011) Synonymous but not the same: the causes and consequences of codon bias. Nat Rev 12:32–42. https://doi.org/10.1038/nrg2899
Prugnolle F et al (2010) African great apes are natural hosts of multiple related malaria species, including Plasmodium falciparum. Proceed Natl Acad Sci USA 107:1458–1463. https://doi.org/10.1073/pnas.0914440107
Radó-Trilla N, Albà M (2012) Dissecting the role of low-complexity regions in the evolution of vertebrate proteins. BMC Evolut Biol 12:155. https://doi.org/10.1186/1471-2148-12-155
Rich SM, Xu G (2011) Resolving the phylogeny of malaria parasites. Proceed Natl Acad Sci USA 108:12973–12974. https://doi.org/10.1073/pnas.1110141108
Saitou N (2018) Introduction to evolutionary genomics. Springer. https://doi.org/10.1007/978-1-4471-5304-7
Salichs E et al (2009) Genome-wide analysis of histidine repeats reveals their role in the localization of human proteins to the nuclear speckles compartment. PLoS Genet. https://doi.org/10.1371/journal.pgen.1000397
Sato S (2021) Plasmodium-a brief introduction to the parasites causing human malaria and their basic biology. J Physiol Anthropol 40:1. https://doi.org/10.1186/s40101-020-00251-9
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27(3):379–423. https://doi.org/10.1002/j.1538-7305.1948.tb01338.x.hdl:10338.dmlcz/101429
Sharp PM, Li WH (1987) The codon adaptation index–a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Research 15:1281–1295. https://doi.org/10.1093/nar/15.3.1281
Shen H, Kan JL, Green MR (2004) Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Molecular Cell 13:367–376. https://doi.org/10.1016/s1097-2765(04)00025-5
Strachan T, Goodship J, Chinnery P (2019) Genetica e genomica nelle scienze mediche. Zanichelli
Sueoka N (1995) Intrastrand parity rules of DNA base composition and usage biases of synonymous codons. J Mol Evol 40:318–325. https://doi.org/10.1007/BF00163236
Sueoka N, Kawanishi Y (2000) DNA G+C content of the third codon position and codon usage biases of human genes. Gene 261:53–62. https://doi.org/10.1016/s0378-1119(00)00480-7
Sun X, Yang Q, Xia X (2013) An improved implementation of effective number of codons (nc). Mol Biol Evol 30:191–196. https://doi.org/10.1093/molbev/mss201
Tachibana S et al (2012) Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat Genet 44:1051–1055. https://doi.org/10.1038/ng.2375
Toll-Riera M et al (2012) Role of low-complexity sequences in the formation of novel protein coding sequences. Mol Biol ution 29:883–886. https://doi.org/10.1093/molbev/msr263
Valkiunas G (2000) Avian malaria parasites and other haemosporidia. CRC Press, NW Corporate Blvd., Boca Raton, Florida
Valkiunas G et al (2018) Characterization of Plasmodium relictum, a cosmopolitan agent of avian malaria. https://doi.org/10.1186/s12936-018-2325-2
Verstrepen KJ et al (2005) Intragenic tandem repeats generate functional variability. Nature Genet 37:986–990. https://doi.org/10.1038/ng1618
Videvall E (2018) Plasmodium parasites of birds have the most AT-rich genes of eukaryotes. Microbial Genom. https://doi.org/10.1099/mgen.0.000150
Waters AP, Higgins DG, McCutchan TF (1993) Evolutionary relatedness of some primate models of Plasmodium. Mol Biol Evol 10:914–923. https://doi.org/10.1093/oxfordjournals.molbev.a040038
Waters AP, Higgins DG, McCutchan TF (1993) The phylogeny of malaria: a useful study. Parasitol Today (Personal ed.) 9:246–250. https://doi.org/10.1016/0169-4758(93)90066-o
Wootton JC, Federhen S (1996) Analysis of compositionally biased regions in sequence databases. Methods Enzymol 266:554–571. https://doi.org/10.1016/s0076-6879(96)66035-2
Wright F (1990) The “effective number of codons” used in a gene. Gene 87:23–29. https://doi.org/10.1016/0378-1119(90)90491-9
Xue HY, Forsdyke DR (2003) Low-complexity segments in Plasmodium falciparum proteins are primarily nucleic acid level adaptations. Mol Biochem Parasitol 128:21–32. https://doi.org/10.1016/s0166-6851(03)00039-2
Yadav MK, Swati D (2012) Comparative genome analysis of six malarial parasites using codon usage bias based tools. Bioinformation 8:1230–1239. https://doi.org/10.6026/97320630081230
Acknowledgements
The authors want to thank the anonymous referees whose corrections and questions have allowed us to greatly improve the work. Furthermore, we thank our Editor, prof. Horacio Naveira, for the extraordinary editing service and management of the entire peer review process.
Author information
Authors and Affiliations
Contributions
AC and AG conceived the study. AC conducted the analyses and conceived the original manuscript. AC, SF and AG read, edited, and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict or competing of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cappannini, A., Forcelloni, S. & Giansanti, A. Evolutionary pressures and codon bias in low complexity regions of plasmodia. Genetica 149, 217–237 (2021). https://doi.org/10.1007/s10709-021-00126-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-021-00126-6