Abstract
The high lignin content of the seed hull of oilseed rape leads to its black seed color compared to yellow seeded soybean. Reducing lignin and increasing oil and protein content is an important breeding aim in oilseed rape. The objective of the present study was to analyze the inheritance of two rapeseed genetic resources with reduced lignin content in the seeds and to elucidate the effects on seed germination and composition. These resources were crossed to black seeded Express 617 and F1 plants were used to develop two doubled haploid (DH) populations. The two DH populations were tested in field experiments in three to five environments. Significant genetic variation for lignin content and bimodal frequency distributions were found in both populations. SNP marker segregation in contrasting bulks revealed for both populations overlapping narrow genomic regions on chromosome C03, responsible for reduced lignin content. The low lignin bulks had a significantly higher percentage of seeds showing bacterial and fungal growth, but germination and vigor was not affected. Non-targeted metabolome analysis of immature green seeds showed exclusively a depletion of metabolites of the proanthocyanidin pathway in DH lines with a reduced lignin content. In these DH lines, a cinnamate-4-hydroxylase gene copy was identified as candidate gene in the overlapping genomic region of both populations. The newly identified QTL for reduced lignin content on chromosome C03 is valuable for studying additive and epistatic effects in combination with other low lignin genotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Seeds of oilseed rape (Brassica napus L.) are a source of high-quality edible oil and of protein rich meal, which is used for livestock feeding. Considering the nutritional value, the meal of oilseed rape has an excellent balanced composition of essential amino acids (Tan et al. 2011). However, quality and digestibility of the meal is negatively influenced by the presence of high amounts of fiber in the seeds. According to the detergent system developed by Van Soest et al. (1991) three different fiber fractions are distinguished: neutral detergent fiber (NDF), acid detergent fiber (ADF) and acid detergent lignin (ADL). NDF mainly consists of cellulose, hemicellulose and lignin, ADF of cellulose and lignin, and ADL of lignin only. Through calculation of differences between these fractions, contents of cellulose and hemicellulose can be estimated. Traditional oilseed rape has a brown or black seed color, which is associated with high lignin contents in the seed hull (Qu et al. 2013; Carré et al. 2016). Animal nutritionists suggest that ADL is the most nutritionally relevant fiber fraction. It reduces the digestibility and the energy uptake both in ruminant and in monogastric animals (Kracht et al. 2004). Seed color mutants with a reduced ADL content were either identified within the oilseed rape gene pool or they were introduced through interspecific hybridization (Rashid et al. 1994; Rahman 2001; Rahman and McVetty 2011 and references therein). Yellow seed character has been reported to be associated not only with lower dietary fiber content but also with higher oil and protein content (Stringham et al. 1974; Simbaya et al. 1995; Rahman et al. 2001). During the past 30 years, developing yellow seeded oilseed rape has become an important breeding objective (Rahman and McVetty 2011). However, despite intensive breeding efforts and reported higher seed oil and protein contents of yellow seeded oilseed rape, it has not yet been possible to establish competitive yellow seeded oilseed rape cultivars in the market. Correlations of ADL content to seed coat phenolic compounds suggest that low ADL content is associated with reduced seed coat thickness (Wittkop et al. 2012; Behnke et al. 2018). Multiple genes and loci have been reported to be involved in the inheritance of seed coat color and testa thickness not only in Arabidopsis thaliana but also in Brassica species (e.g. Badani et al. 2006; Stein et al. 2013; Yan et al. 2009; Liu et al. 2015; Taylor-Teeples et al. 2015; Wang et al. 2015, 2017; Körber et al. 2016).
The seed coat controls seed germination and can be seen as a barrier for water and/or oxygen, or by providing mechanical resistance to radicle protrusion. In Arabidopsis, inhibited germination had been positively correlated with seed coat color, due to the presence of phenolic compounds (Debeaujon et al. 2000). In spite of all reported advantages of yellow seed coat, the thinner testa in yellow-seeded types also means the seed is more prone to damage by various environmental factors (Zhou et al. 2020). Seeds of oilseed rape are reported to maintain germination capacity for 10 years and longer in the soil. Thereby germination is prevented by secondary induced dormancy until seeds may finally germinate under favorable conditions in the soil (Schatzki et al. 2013 and references therein). Seed longevity declines also during dry storage (Rajjou and Debeaujon 2008) and the ageing rate depends on seed moisture content, temperature, initial seed quality and on genetic factors (Nagel et al. 2011, 2018). Seed longevity of yellow-seeded types tends to drop more rapidly compared to the black-seeded ones (Debeaujon et al. 2003). Since materials of naturally aged seeds are not always available, artificial seed ageing protocols are often utilized. Exposure of seeds to high temperature and high moisture conditions has been a commonly used method for ageing seeds in the laboratory (Hay et al. 2019).
In different populations a large number of QTL on linkage groups A05, A06, A09, C05, C08, C09, for reduced lignin content and yellow seed color have been published (Badani et al. 2006; Stein et al. 2013; Wang et al. 2015, 2017; Behnke et al. 2018; Miao et al. 2019). Despite the identification of numerous loci affecting seed coat color in the allopolyploid genome of oilseed rape, none of the identified QTL have led to the development of competitive true yellow seeded rapeseed cultivars. It has been argued that loci for yellow seed character may have negative pleiotropic effects on seed germination, pre-harvest germination, seed longevity and vigor. The objective of this work was to study the inheritance of the yellow seed character in relation to seed germination, seed longevity, pre-harvest germination and other seed quality traits. For this purpose, two not yet characterized yellow seeded genetic resources were crossed to the same black seeded inbred line and the derived two doubled haploid (DH) populations were characterized following field experiments.
Material and methods
Plant material
Population I consisted of 75 DH lines derived from the cross of genotype 4042 × Express 617 and the two parents. Line 4042 is a winter oilseed rape of the Department of Crop Sciences at the University of Göttingen. Seeds of 4042 have a yellowish-brown appearance and a reduced content of Acid Detergent Lignin (ADL). They are high in glucosinolate content but have low erucic acid content in the seed oil. Express 617 is a black seeded inbred line of the German winter oilseed rape cultivar Express and seeds are of canola quality. Population II consisted of 135 DH lines derived from the cross DH1372 × Express 617. DH1372 is a yellow-seeded canola (‘00’) DH line derived from the cross between the two black seeded Canadian spring cultivars Star and Bolero (Burbulis and Kott 2005). Seed material of DH1372 was provided by Laima Kott (University of Guelph). Both DH populations were developed at the University of Göttingen. Since DH1372 is a spring genotype it could not survive winter in field trials performed in north-western Germany.
Field experiments
Population I was tested in field experiments in five environments, in Göttingen-Reinshof and Einbeck, both located in north-western Germany in 2014, 2015 and 2016. Population II was tested in Göttingen-Reinshof in the three subsequent years 2015, 2016 and 2017. One hundred seeds were sown in single row observation plots. Fertilizer, fungicides and insecticides were applied following standard schemes at each location. At maturity, seeds were harvested and bulked from 10 main racemes of open pollinated plants from each plot. Seeds were dried at 30 °C to a moisture content of 5 to 8% and stored at a dry place at room temperature for further analysis. From population II 224 DH lines were sown in 2015. Due to segregation of the spring rapeseed character and unfavorable weather conditions during winter 2015/16 seeds from only 135 DH lines were harvested.
Seed quality analysis
Near Infrared Reflectance Spectroscopy (NIRS) was applied to predict moisture, oil, protein, and glucosinolate content of the seed, and fiber contents of the defatted meal (NDFm, ADFm, and ADLm). NIRS measurements were performed using seed samples of 3 g in small ring cups and the FOSS monochromator model 6500 (NIRSystem Inc., Silverspring, MD, USA). Oil (%), protein (%), and glucosinolate (µmol/g seed) contents were predicted at 91% seed dry matter content using the commercial calibration equations of raps 2012.eqa provided by VDLUFA Qualitätssicherung NIRS GmbH (Am Versuchsfeld 13, D-34128 Kassel, Germany). Total seed oil and protein content (oil + P) was obtained by forming the sum of oil and protein content. The values of oil and protein content were further used to calculate protein content of defatted meal (PDM): PDM (%) = [% protein/(100% oil)] × 100. NDFm, ADFm and ADLm contents were determined by calibration equations fibr2013.eqa (Dimov et al. 2012). All fiber content values are given as percentage of fiber in the defatted meal.
Determination of other seed traits
Thousand seed weight (TSW) was calculated by taking the weight of 500 seeds. Percentage of pre-harvest germination (PHG) was determined by counting the number of seeds showing pre-harvest germination in a sample of 100 randomly selected seeds. Seed color was visually scored according to a scale from (3) for yellow brown, (5) for brown, (7) for dark brown and (9) for black and also using intermediate scores. To avoid effects caused by primary seed dormancy, seed germination was performed with seeds that have been stored for 6 weeks after field harvest. The germination test was carried out in Petri dishes (92 × 16 mm, Sarstedt, reference code 82.1473) using customized filter paper (90 mm in diameter, Macherey-Nagel, GmbH & Co. KG, reference code 400,866,009.1) with 50 indented holes each to hold seeds. Fifty randomly chosen intact seeds per DH line and Petri dish were tested. Mean values for 100 seeds were used in statistical analysis of seed germination. Twelve milliliters of de-ionized water were carefully added. Petri dishes were covered with pertinent coverlids and they were placed in large plastic trays covered with cellophane to reduce evaporation. The trays were placed in darkness in a germination chamber at a temperature 16.5–17.5 °C. After 10 days, seeds were inspected and radicle protrusion (%), germination (%), contamination with bacteria or fungi (%) and hypocotyl length were determined. Germination was scored when the radicle was elongated (> 1 cm) and both cotyledons were outside of the seed coat. Radicle protrusion was scored when seed radicle elongated visually and protruded out of seed coat, but the cotyledons were not yet swollen and were still embedded within the seed coat. Contaminated seeds were identified by bacterial/fungal growth around seeds on filter paper. Based on slime or mycelium formation around the seeds, contaminations were considered to be caused by bacteria or fungi, respectively. Furthermore, average hypocotyl length in cm was measured as a proxy for seedling vigor. Seed germination after artificial ageing was determined following the ‘controlled deterioration’ procedure of Hay et al. (2008). Seeds were equilibrated above 8.7 molar non-saturated lithium chloride (LiCl) solution at 20 °C to reach 47% relative humidity (RH) for 10 days. Then, seeds were placed above 7.1 molar LiCl solution at 45 °C to increase the RH to 60% and were kept for 50 days. Afterwards, two replicates of 50 seeds each were germinated for 10 days in Petri dishes under the same conditions and were inspected for the same traits as described above.
Non-targeted metabolite analysis of segregating bulks of the population 4042 × Express 617
Non-targeted metabolite finger printing according to Feussner and Feussner (2019) was applied to detect differences in the metabolite abundance between yellow and black seeded DH lines of population I. Fully mature dry seeds derived from DH lines with contrasting low and high ADLm contents were analyzed. However, no clear differences in relative metabolite abundance between the two groups were found. Therefore, each 15 DH lines with low and with high ADLm content in mature seeds were sown in the greenhouse. At BBCH plant developmental stage 78 to 79 (fully grown siliques with green seeds; three to four weeks after self-pollination; Meier 2001; Hajduch et al. 2006) siliques were harvested and carefully opened with a scalpel. Green seeds were collected and immediately frozen in liquid nitrogen. Frozen seeds of five plants each with low and high ADLm content were bulked to give in total six bulks of seeds (three biological replicates with each low and high ADLm content). In order to check if identical or different changes in relative metabolite abundance occur in DH1372, green seeds of parental genotype DH1372 were also included with three biological replicates in the analysis. 150 seeds were homogenized for each replicate bulk using a Mixer Ball Mill MM200 (Retsch GmbH). For metabolite extraction, 80 mg of the frozen seed material were subjected to an adapted two-phase extraction with methyl-tert-butylether (MTBE), methanol and water (Feussner and Feussner 2019). Metabolite fingerprinting was performed with an UHPLC 1290 Infinity (LC, Agilent Technologies) coupled to a high-resolution mass spectrometer (HR-MS, 6540 UHD Accurate-Mass Q-TOF, Agilent Technologies) with Agilent Dual Jet Stream Technology as electrospray ionization (ESI) source (Agilent Technologies). For chromatographic separation of the samples from the polar as well as the non-polar extraction phase, an ACQUITY HSS T3 column (2.1 × 100 mm, 1.8 μm particle size, Waters Corporation) was used as stationary phase at 40 °C. The solvents A (water with 0.1% (v/v) formic acid) and B (acetonitrile with 0.1% (v/v) formic acid) were used to set-up the following gradient: 0 to 3 min: 1% to 20% B; 3 to 8 min: 20% to 100% B; 8 to 12 min: 100% B; 12 to 15 min: 1% B. The flow rate was set to 500 µl/min. The Q-TOF MS instrument was used in a range from m/z 50 to m/z 1700 with a detection frequency of 4 GHz, a capillary voltage of 3000 V, nozzle and fragmentor voltage of 200 V and 100 V, respectively. The flow of drying gas was set to 8 l/min and sheath gas to 8 l/min, respectively. The sheath gas was set to 300 °C, and drying gas to 250 °C. Data were acquired in positive as well as negative ESI mode with Mass Hunter Acquisition B.03.01 (Agilent Technologies). Profinder B.08.02 (Agilent Technologies) was used to generate data matrixes. The MarVis-Suite toolbox (Kaever et al. 2015, http://marvis.gobics.de/) was used for data processing, statistics, data mining and visualization. First, metabolite features with a false discovery rate (FDR) < 0.005 were selected. Overall, 486 features obtained from positive ESI mode as well as 254 features from negative ESI mode of the polar extraction phase and 225 features from positive ESI mode as well as 104 features from negative ESI mode of the non-polar extraction phase were selected, merged and used for clustering. The accurate mass information of the selected metabolite features was used for metabolite annotation (KEGG, http://www.kegg.jp and BioCyc, http://biocyc.org). Verification of the chemical structure of the annotated metabolite markers was performed by LC-HR-MSMS analyses (Online Resource 1) as described by Feussner and Feussner (2019). A principal component analysis (PCA) was performed with the filtered data set of 1,069 metabolite features (FDR < 0.005) using the software MetaboAnalyst (Chong et al. 2019, https://www.metaboanalyst.ca/).
Bulk segregant SNP-marker analysis
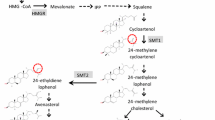
Frequency distributions of seed ADLm content of the DH populations 4042 × Express 617 and DH1372 × Express 617 showed a bimodal type of frequency distribution (Online Resource 2) which suggested segregation of one major gene controlling this trait. A bulk segregant analysis was performed using a proprietary Brassica 20 K Illumina Infinium™ SNP array (KWS SAAT SE, www.kws.de) for population I and the Brassica 15 K Illumina Infinium™ SNP array of TraitGenetics GmbH (www.traitgenetics.com) for population II. For population I, four bulks each consisting of 5 DH lines with low ADLm content and four bulks each consisting of 5 DH lines with high ADLm content were analyzed. For population II, three bulks with low and two bulks with high ADLm content, each consisting of 5 DH lines were analyzed. SNP markers specific for the low and high ADL bulks were identified. Selected SNP markers (c.f. Figure 1) were used to genotype the complete populations applying the KASP marker technology (LGC group; www.lgcgroup.com/genomics). SNP marker sequences were provided by Isobel Parkin (AAFC, Saskatoon, Canada; Clarke et al. 2016) and KASP markers were ordered from the LGC group (www.lgcgroup.com/genomics). KASP marker analysis was performed following suppliers’ instruction in 96 well plates using a CFX96 Touch Real-Time PCR Detection System. SNP sequences were aligned against the Darmor-bzh B. napus reference genome v4.1 (http://www.genoscope.cns.fr/brassicanapus/, Chalhoub et al. 2014) and against the ZS11 reference sequence (https://www.ncbi.nlm.nih.gov/assembly/GCF_000686985.2/, Sun et al. 2017). Most likely positions were selected from the BLAST hits considering best matching and highest possible E-value as well as genetic map data information (Fig. 1). SNP marker positions were also compared with the positions listed in the supplementary material of Clarke et al. (2016) Annotation of the B. napus gene sequences was performed by Boas Pucker based on the Araport11 complete reannotation of the Arabidopsis thaliana reference genome (Cheng et al. 2017).
Physical map with SNP markers on chromosome C03 that allows distinguishing bulks of genotypes with low and high ADL content in the two different DH populations. Physical positions of SNP marker and the cinnamate-4-hydroxylase gene (BnaC03g16950D and BnaC03g16960D) are based on the Darmor-bzh reference genome (Chalhoub et al. 2014)
Statistical analysis
Analysis of variance and prediction of heritability values were performed by PLABSTAT software (Utz 2011). Both environment and genotype factors were considered as random variables. The general model for analysis of variance is as follow:
where Y is observation of genotype i in environment j; µ is general mean; gi and ej were the effects of genotype i and environment j; geij is the interaction between genotype x environment of genotype i with environment j. Broad sense heritability (h2) was calculated as follows: h2 = δ2G/(δ2G + δ2GE/E) where σ2G is the variance component for genotype, σ2GE is the variance component for genotype and environment interaction, and E is the number of environments. Least significant differences (LSD5%) and Spearman’s rank correlation coefficients between trait means were calculated by PLABSTAT software (Utz 2011). Newman Keuls comparisons of contrasting groups were performed with STATISTICA data analysis software system (version13.1; StatSoft, I. N. C. (2016) using mean values over all environments.
Results
DH population of the cross 4042 × Express 617
Significant genotypic variation within this population was detected for all seed quality traits without and with artificial ageing treatment except for seed contamination without artificial ageing treatment (Online Resource 3). Heritabilities ranged from modest for glucosinolate content (0.26) to high for seed fiber (0.81 to 0.86) content and seed color (0.95). Parental line Express 617 had a significantly higher oil content (48.8%) and lower seed protein content (16.9%) compared to line 4042 (40.7 and 18.9, respectively; Table 1). Significant and relatively large differences between the parental lines and within the DH population were found for the seed fiber constituents NDFm, ADFm and ADLm and for seed color. Thereby, line 4042 had significantly lower fiber contents and lower seed color score compared to black seeded Express 617 and a comparable range was also detected in the DH population. Noteworthy, seeds of line 4042 showed much higher contaminations than Express 617. Both parental genotypes had a seed germination above 97%. In the DH population seed germination ranged from 81.5 to 100% with a mean of 98.4%. The artificial ageing treatment led to an increased germination range from 20.8 to 86.2% and a reduction to 62.8% as a mean of the population. This was accompanied by an increase in seeds showing only radical protrusion (i.e. incomplete germination) and a reduction by more than half of hypocotyl length. Furthermore, the artificial ageing treatment increased percentage of seeds with contaminations in the population from 3.2 to 11.0%.
Spearman rank correlation analysis showed that ADLm content was closely positive correlated with ADFm and NDFm content. The three fiber fractions were slightly positive correlated with seed germination (for ADLm rS = 0.21) which became significant after artificial seed ageing (rS = 0.45**). In contrast, all fiber fractions were negatively correlated with pre-harvest germination, radical protrusion and contaminated seeds (Table 2) and similar correlations were obtained following artificial ageing treatment. Notably, the correlation between ADLm content and hypocotyl length became significant positive only after artificial seed ageing.
Frequency distribution of the ADLm content did not show a normal distribution. It was significantly skewed to the right (Skewness 0.46, p = 0.10; Online Resource 2) and suggested a bimodal segregation with a possible separation of two groups at 10% ADLm content. Comparing the two groups separated at 10.0% ADLm content confirmed significant differences between the two groups for seeds with contaminations and for all traits recorded following artificial seed ageing (Table 3). Separation of the two groups also showed that a larger number of DH lines with a low ADLm content regenerated from microspore culture in the DH population than DH lines with a high ADLm content (Table 3). The clear differences between the low and high ADLm bulks and the bimodal segregation of ADLm content in the DH population suggested that the difference might be caused by one major QTL. Bulk segregant analysis of DH lines with low and high ADLm content using the 20 K Illumina SNP-chip allowed to locate bulk specific SNP markers on chromosome C03 (Online Resource 4; Fig. 1). Three SNP-markers were selected and KASP-marker analysis was performed on the complete population of 75 DH lines. With some overlap at 10% ADLm content, the KASP markers allowed unequivocally distinguishing DH lines of the low and high ADLm group (data not shown).
DH population of the cross DH1372 × Express 617
Seed quality analysis revealed significant effects for the genotype and the environment for most of the traits of the DH1372 × Express 617 DH population (Online Resource 3). Heritabilities ranged from 0.22 for pre-harvest germination to 0.86 for seed color. Compared to the 4042 × Express 617 DH population, relatively low heritabilities were obtained because experiments were performed only in three environments. As for the first DH population, the frequency distribution of the ADLm content of DH1372 × Express 617 DH population was significantly skewed to the right (Skewness 0.51, p = 0.05; Online Resource 2). Again, the distribution suggested a bimodal segregation with a possible separation of two groups at 9.0% ADLm content. Separating the two groups at 9% ADLm content showed again a much larger group of DH lines with low ADLm content (n = 94) compared to the group with high ADLm content (n = 41). Seeds of the two different groups of DH lines were further analyzed for their germination without and with artificial ageing (Table 4). The low ADLm group had significantly higher contents of oil and of the sum of oil and protein. Furthermore, the low ADLm group had without and with artificial ageing significantly higher percentage of contaminated seeds. After the artificial ageing treatment, hypocotyl length decreased from 4 cm to less than half after 10 days of germination. As for population 4042 × Express 617, seeds with low ADLm content had significantly shorter hypocotyls after artificial ageing treatment. However, compared to population 4042 × Express 617, the seed ageing treatment for 50 days did not affect germination of DH1372 x Express 617. There was also a lack of correlations between fiber contents and seed germination after artificial ageing (Table 5). Bulk segregant analysis of DH lines with low and high ADLm content using the 15 K Illumina SNP-chip allowed to locate bulk specific SNP markers on chromosome C03 (Online Resource 4; Fig. 1). Surprisingly, the identified region was overlapping with the region identified in population 4042 × Express 617. Six SNP-markers were selected and KASP-marker analysis was performed on the whole population. With some overlap at 9% ADLm content, the KASP markers allowed unequivocally distinguishing DH lines of the low and high ADLm group (data not shown).
Non-targeted metabolite analysis of the DH population 4042 × Express 617
Non-targeted metabolome analysis of green seeds of the contrasting bulks for high and low ADLm content of the DH population 4042 × Express 617 and the low ADLm genotype DH1372 resulted in 1069 significantly different metabolite features (FDR < 0.005). The intensity profiles of these features were clustered and visualized by means of 1D-SOMs (one-dimensional self-organizing maps, feature based clustering) and organized into six clusters (Online Resource 5). Cluster 1 to 3 represent 732 metabolite features which are characterized by higher abundance in genotype DH1372 in comparison to both bulks of the DH population 4042 × Express 617. Features (279), which are summarized in cluster 4 and 5 show the inverse pattern with higher abundance in both bulks of the DH lines (low and high ADLm content). Cluster 6 contains 58 metabolite features that show high intensities in the high ADLm bulk in contrast to the low ADLm bulk and the low ADLm genotype DH1372. Since LC-ESI-MS analyses tend to form a considerable number of adducts, the 58 low ADLm specific features may represent in total about 15 metabolites. Low ADLm genotype DH1372 displayed a completely different metabolite feature in comparison to the 4042 × Express 617 low and high ADL bulks. This is not surprising, because genotype DH1372 is a Canadian spring DH-line that is genetically distant to European winter oilseed rape material. However, DH1372 showed a similar low metabolite intensity in cluster 6 as the low ADL bulk of 4042 × Express 617. The accurate mass of the metabolite features represented in cluster 6 was used for an automated data base search (against BioCyc and KEGG) to assign tentative identities to the features. Exclusively metabolites of the procyanidin biosynthesis were identified as enriched in the high ADLm bulk. The identity of leucocyanidin, catechin and the procyanidin-oligomers including two to five procyanidin units were unequivocally confirmed by LC-HR-MSMS analysis (Online Resource 5). Other precursors of the procyanidin pathway like phenylalanine, cinnamic acid, coumaroyl-CoA, chalcone or flavanones could not be detected among the metabolites in cluster 6. Sample-based clustering of the data set by a PCA showed a clear separation on principal component 1 (PC1) between the Canadian spring genotype DH1372 and the European winter oilseed rape material (4042 × Express 617), while on PC2 all three samples groups (DH1372, the low and the high 4042 × Express 617 bulks) were separated (Online Resource 6).
Identification of candidate genes
Bulk segregant analysis allowed confining the physical region responsible for reduced ADLm content on chromosome C03 in both populations. Using the SNP-marker sequences, the physical region between the first and last marker were characterized. In the Darmor-bzh reference genome the physical position of the Bn-scaff_21778_1-p215559 marker was at 5.012.004 bp and that of the to the last marker Bn-scaff_21312_1-p1309658 was at 9.145.606 bp (Fig. 1). In the ZS11 reference genome this region spanned from 10.393.115 to 18.397.417 bp. The physical region between the two flanking markers of the Darmor-bzh (Online Resource 7) and the ZS11 reference genome sequence (Online Resource 8) was inspected for candidate genes known to be involved in the phenylpropanoid and lignin biosynthetic pathway. Candidate genes comprised a number of Myb transcription factors (myb14, myb19, myb82, myb49, myb120, myb36, myb119, myb59) and a cinnamate-4-hydroxylase gene (Fig. 1) that are marked yellow in Online Resources 7 and 8. The most likely candidate gene is the cinnamate-4-hydroxylase gene at position 8.615.302 bp in Darmor-bzh and at position 17.639.366 in ZS11. In both reference genomes, this gene is present in tandem duplicate. Although the two genes in Darmor-bzh (BnaC03g16950D and BnaC03g16960D) are outside flanking marker Bn-scaff_18322_1-P1044275, the distance to this marker is less than 600 kb. In ZS11 the two genes BnC03g0556270.1 and BnC03g0556290.1 are located well within the region delimited by the group specific SNP-markers. BLAST of the ZS11 gene sequences against the Darmor-bzh genome showed that the genes corresponded to each other in the same order. A MATE efflux family protein (BnaC03g18950D) was identified in the Darmor-bzh genome a bit outside the group specific marker range at position 9.813.528. In the ZS11 genome the MATE efflux family protein gene BnC03g0554010.1 was identified at 15.788.740 bp. A little outside the flanking markers in Darmor-bzh at 10.964.534 bp and in ZS11 at 19.126.045 bp the transparent testa gene BnaC03g20650D is located.
Discussion
Development of oilseed rape cultivars with a reduced fiber content and increased oil and protein content is a desirable breeding objective. Genetic variation in oilseed rape may affect seed hull proportion and phenylpropanoid content of the seed hull. Distribution of the fiber components is unbalanced in the seed. Fiber content is especially high in seed hull which contains around 70% of NDF, 80% of ADF and more than 90% of the lignin (ADLm) content of the seed (Carré et al. 2016). Through technical dehulling of seeds, protein content in defatted meal can be increased from about 38% to 48% (Carré et al. 2016).
Despite existing genetic variation for seed fiber content, breeding of competitive yellow seeded oilseed rape cultivars has not yet been successful. Disadvantages of seeds with lower fiber content may include lower germination, increased pre-harvest germination, reduced seed longevity and vigor (Zhou et al. 2020). These traits negatively affect the crops performance in the field. Furthermore, a thinner seed hull may lead to damaged seeds by combined harvesting which in turn may reduce oil quality by increasing free fatty acid content. Instead of looking for major genes causing drastic reduction of seed hull and phenylpropanoid content, breeders may combine different minor loci for reduced fiber content to develop breeding lines, which do not show any of the mentioned disadvantages. Detecting effects of minor loci and combining them for reduced fiber content can be quite cumbersome and time consuming. However, breeding becomes easy, once a number of alleles for reduced fiber content are fixed in breeding material. Surprisingly, no negative pleiotropic effects of the bright yellow seed color of Yellow Sarson (B. rapa) on pre-harvest germination and seed longevity has been reported. Notably, Bagheri et al. (2012) mapped in a RIL population derived from a cross between a black seeded B. rapa line and the yellow seeded B rapa Yellow Sarson a major QTL for yellow seed color on chromosome A09 explaining 54% of the phenotypic variation. Furthermore, Bagheri et al. (2012) also investigated in the same RIL population the inheritance of pre-harvest germination (vivipary) and found no correlation to the yellow seed character in the population. Furthermore, although the largest QTL for pre-harvest germination was identified at the same chromosome A09, it mapped more than 50 cM away. Together, this indicates that at least for this locus there is no negative pleiotropic effect of the yellow seed color alleles on pre-harvest germination. Previously, another major QTL for preharvest germination was identified on chromosome N11 of B. napus (Feng et al. (2009). In the present work only a slight negative and no correlation between ADLm content and pre-harvest germination was found for the first and second population, respectively, which is in support of an independent inheritance of this trait from reduced lignin content. Interestingly, Bagheri et al. (2012) discussed that the major QTL for yellow seed color on chromosome A09 of the B. rapa population maybe the same as a QTL on A09 causing a reduced ADL content in a B. napus DH population (Liu et al. 2012; Stein et al. 2013). It was suggested that a Cinnamoyl-CoA Reductase (CCR1) locus may be responsible for this effect. CCR1 catalyzes in the phenylpropanoid pathway the step from p-coumaric acid to 4-coumaroyl-CoA. It would be interesting to test whether resynthesized B. napus lines derived from crosses between Yellow Sarson and black seeded B. oleracea genotypes (c.f. Jesske et al. 2013) would show reduced lignin contents in the seed hull.
In both DH populations in very similar regions a QTL was identified on chromosome C03. It remains unclear, if the two QTL were caused by mutations in the same or different genes. Since, one genotype was derived from Canadian spring canola material (Burbulis and Kott 2005) and the other genotype was a Göttinger resource with high glucosinolate content, it appears unlikely that the same genes were affected in both genotypes. Assuming that the QTL in both genotypes is located at a very similar position it would require a large segregating F2 population to separate the two loci. Reciprocal crossing of both homozygous genotypes to produce F1 seeds is also not conducive, because seed hull is maternal tissue. Assuming the genetic causes for low lignin content are different in DH1372 and 4042, one could estimate additive and synergistic effects, if the composition of the seed hull were determined by the genotype of the embryo.
There was no difference in metabolite content when fully mature seeds of the low and high ADLm groups were analyzed by non-targeted metabolome analysis. However, analyzing immature green seeds revealed exclusively metabolites of procyanidin biosynthesis were strongly depleted in the low ADLm 4042 × Express 617 group (Online Resource 5). Akhov et al. (2009) and Miao et al. (2019) obtained comparable results when they analyzed transcript abundance by qRT-PCR experiments. They found gene expression differences in early seed developmental stages but not in mature seeds. Despite gross differences in clusters 1 to 5 of yellow seeded DH1372 in comparison to the 4042 × Express 617 groups, DH1372 showed an equal reduction of procyanidin metabolites as the low ADLm 4042 × Express 617 group. The likewise reduction of the same procyanidin metabolites in the low ADLm group of 4042 × Express 617 and in DH1372 suggests that in both genotypes genes of the procyanidin pathway could be affected. Hong et al. (2017) found a number of biosynthetic and regulatory genes in seed coats, whose down-regulation mainly contributed to the reduction of procyanidins in yellow seeds. Similarly, Qu et al. (2020) found in a comparison of different yellow and black seeded genotypes, that genes of the procyanidin pathway had lower expression levels in yellow-seeded rapeseed. Based on metabolite profiles they suggested that the seed coat color could be mainly determined by the levels of epicatechin and their derivatives. In genotypes with mutations in different genes of the procyanidin pathway, the phenotypic effect on the metabolite composition may be different. Considering the large number of possible candidate genes in the QTL region for the observed phenotypic differences, non-targeted metabolite analysis could represent a valuable additional tool to distinguish between genotypes with different mutations (Qu et al. 2020). Within the genomic region delimited by the flanking SNP markers in the Darmor-bzh and the ZS11 reference genome a number of candidate genes were identified. A likely candidate is the cinnamate-4-hydroxylase—gene that occurs in this region in tandem duplicate in both reference genomes. Cinnamate-4-hydroxylase catalyzes the conversion of cinnamic acid to p-coumaric acid, the second step in the general phenylpropanoid pathway (Liu et al. 2015; Hong et al. 2017). Since above mentioned CCR1 catalyzes in the phenylpropanoid pathway the next step from p-coumaric acid to 4-coumaroyl-CoA, it is clear that the same pathway is affected by mutations. An impaired activity of Cinnamate-4-hydroxylase could very well lead to reduced contents of downstream metabolites of this pathway. None of the myb transcription factor genes detected within the flanking markers were so far reported to be activators or repressors of the phenylpropanoid pathway (Liu et al. 2015; Wei et al. 2017; Hong et al. 2017). Candidate gene BnaC03g18950D is a transporter protein, also known as multidrug and toxin extrusion (MATE) protein. In Arabidopsis, AtTT12 encodes a transporter protein that acts as a proton-dependent antiporter, assisting vacuolar localization of proanthocyanidins in the testa (Debaujon et al. 2001). Transparent Testa (TT12) transporter proteins have been reported to be involved in yellow seed character in Arabidopsis and Brassica species (Chai et al. 2009; Hong et al. 2017). Similarly, candidate gene BnaC03g20650D is a WRKY family transcription factor protein, which is also known as BnTTG2 transparent testa gene (Johnson et al. 2002; Qu et al. 2013; Liu et al. 2016, Qu et al. 2020).
Although both DH populations showed similar large and significant differences in ADLm, ADFm and NDFm content between the low and high ADLm groups, only DH population DH1372 × Express 617 showed a significant increase in oil content and in the sum of oil and protein content. This was not noticed in population 4042 × Express617, albeit this population was tested more thoroughly in five environments. This illustrates that positive effects of reduced lignin content alleles on oil and protein content may depend either on the genetic background or on the affected lignin type locus (Taylor-Teeples et al. 2015). In both DH populations there were only non-significant minor differences in traits like pre-harvest germination, thousand seed weight and hypocotyl length. To date only few studies have been performed on analyzing the effect of reduced seed fiber content on seed germination traits. In both DH populations there were no significant differences in seed germination of the low and high ADLm group (Tables 3 and 4). The artificial seed ageing treatment reduced in both populations the seed germination, but only in DH population 4042 × Express 617 the low ADLm group showed a comparatively stronger reduction in germination after artificial seed ageing treatment. This result is in contrast to a lower germination percentage and lower seed vigor index after artificial ageing of near-isogenic yellow-seeded and black-seeded lines as reported by Zhang et al. (2006). However, in the present study the low ADLm group of both populations had a 5 to 7% higher percentage of seeds showing contaminations (Tables 3 and 4). This difference was also observed after artificial seed ageing. Interestingly, the higher contamination rate of the low ADLm group had little effect on seed germination in both populations. Only the seed vigor, i.e. the hypocotyl length of both populations was reduced in the low ADLm group that may have been additionally influenced by the contamination. The microbial species of the seed contaminations has not been determined in the present study, but earlier studies have shown that seed born Alternaria spores may affect seed quality and germination (Meena et al. 2016; Soomro et al. 2020).
With the result of the present study, a new QTL for reduced lignin content on oilseed rape chromosome C03 has been identified in two different populations. It remains open whether the same gene or different closely linked genes are affected. The plant material can be used in crosses with already identified other lignin mutant genotypes (e.g. Behnke et al. 2018) that carry mutations on different chromosomes of the rapeseed A- or C-genome to identify additive and epistatic effects on lignin content and seed color. Despite many attempts to develop yellow seeded rapeseed, black seeded cultivars are still dominating the market. This is in clear contrast to soybean, where yellow seeded types are representing the commodity and dark seeded types are mainly used as vegetables. As a leguminous species, soybean has the advantage that is has a higher protein content and a lower oil and fiber content. The lower total fiber and especially the lower lignin content of the yellow seeded soybean contributes to the excellent quality of the meal and isolated proteins.
References
Akhov L, Ashe P, Tan Y, Datla R, Selvaraj G (2009) Proanthocyanidin biosynthesis in the seed coat of yellow-seeded, canola quality Brassica napus YN01-429 is constrained at the committed step catalyzed by dihydroflavonol 4-reductase. Botany 87:616–625
Badani AG, Snowdon RJ, Wittkop B, Lipsa FD, Baetzel R, Horn R, De Haro A, Font R, Lühs W, Friedt W (2006) Colocalization of a partially dominant gene for yellow seed colour with a major QTL influencing acid detergent fibre (ADF) content in different crosses of oilseed rape (Brassica napus). Genome 49:1499–1509
Bagheri H, El-Soda M, van Oorschot I, Hanhart C, Bonnema G, Jansen-van den Bosch T, Mank R, Keurentjes J, Meng L, Wu J, Koornneef M (2012) Genetic analysis of morphological traits in a new, versatile, rapid-cycling Brassica rapa recombinant inbred line population. Front Plant Sci 3:183
Behnke N, Suprianto E, Möllers C (2018) A major QTL on chromosome C05 significantly reduces acid detergent lignin (ADL) content and increases seed oil and protein content in oilseed rape (Brassica napus L.). Theor Appl Genet 131:2477–2492
Burbulis N, Kott LS (2005) A new yellow-seeded canola genotype originating from double low black-seeded Brassica napus cultivars. Can J Plant Sci 85:109–114
Carré P, Citeau M, Robin G, Estorges M (2016) Hull content and chemical composition of whole seeds, hulls and germs in cultivars of rapeseed (Brassica napus). OCL 23:A302
Chai YR, Lei B, Huang HL, Li JN, Yin JM, Tang ZL, Wang R, Chen L (2009) TRANSPARENT TESTA 12 genes from Brassica napus and parental species: cloning, evolution, and differential involvement in yellow seed trait. Mol Genet Genom MGG 281:109–123. https://doi.org/10.1007/s00438-008-0399-1
Chalhoub B, Denoeud F, Liu S et al (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345:950–953. https://doi.org/10.1126/science.1253435
Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD (2017) Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89:789–804. https://doi.org/10.1111/tpj.13415
Chong J, Wishart DS, Xia J (2019) Using metaboanalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinform 68:e86
Clarke WE, Higgins EE, Plieske J, Wieseke R, Sidebottom C, Khedikar Y, Batley J, Edwards D, Li R, Lawley CT, Pauquet J, Laga B, Cheung W, Iniquez-Luy F, Dyrszka E, Rae S, Stich B, Snowdon RJ, Sharpe AG, Ganal MW, Parkin IAP (2016) A high-density SNP genotyping array for Brassica napus and its ancestral diploid species based on optimised selection of single-locus markers in the allotetraploid genome. Theor Appl Genet 129:1887–1899. https://doi.org/10.1007/s00122-016-2746-7
Debeaujon I, Léon-Kloosterziel KM, Koornneef M (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122:403–414
Dimov Z, Suprianto E, Hermann F, Möllers C (2012) Genetic variation for seed hull and fibre content in a collection of European winter oilseed rape material (Brassica napus L.) and development of NIRS calibrations. Plant breed 131(3):361–368
Debeaujon I, Peeters AJM, Léon-Kloosterziel KM (2001) The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter-like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13:853–872. https://doi.org/10.1105/tpc.13.4.853
Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L (2003) Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell 15:2514–2531. https://doi.org/10.1105/tpc.014043
Feng F, Liu P, Hong D, Yang G (2009) A major QTL associated with preharvest sprouting in rapeseed (Brassica napus L.). Euphytica 169:57–68
Feussner K, Feussner I (2019) Comprehensive LC-MS-based metabolite fingerprinting approach for plant and fungal-derived samples. High-throughput metabolomics. Humana, New York, pp 167–185
Hajduch M, Casteel JE, Hurrelmeyer KE, Zhao S, Agrawal GK, Thelen JJ (2006) Proteomic analysis of seed filling in Brassica napus. Developmental characterization of metabolic isozymes using high-resolution two-dimensional gel electrophoresis. Plant Physiol 141:32–46. https://doi.org/10.1104/pp.105.075390
Hay FR, Adams J, Manger K, Probert R (2008) The use of non-saturated lithium chloride solutions for experimental control of seed water content. Seed Sci Technol 36:737–746
Hay FR, Valdez R, Lee JS, Sta Cruz PC (2019) Seed longevity phenotyping: recommendations on research methodology. J Exp Bot 70:425–434
Hong M, Hu K, Tian T, Li X, Chen L, Zhang Y, Yi B, Wen J, Ma C, Shen J, Fu T (2017) Transcriptomic analysis of seed coats in yellow-seeded Brassica napus reveals novel genes that influence proanthocyanidin biosynthesis. Front Plant Sci 8:1674
Jesske T, Olberg B, Schierholt A, Becker HC (2013) Resynthesized lines from domesticated and wild Brassica taxa and their hybrids with B. napus L.: genetic diversity and hybrid yield. Theor Appl Genet 126:1053–1065
Johnson CS, Kolevski B, Smyth DR (2002) TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell 14:1359–1375
Kaever A, Landesfeind M, Feussner K, Mosblech A, Heilmann I, Morgenstern B, Feussner I, Meinicke P (2015) MarVis-Pathway: integrative and exploratory pathway analysis of non-targeted metabolomics data. Metabolomics 11:764–777
Körber N, Bus A, Li J, Parkin IAP, Wittkop B, Snowdon RJ, Stich B (2016) Agronomic and seed quality traits dissected by genome-wide association mapping in Brassica napus. Front Plant Sci 7:386. https://doi.org/10.3389/fpls.2016.00386
Kracht W, Danicke S, Kluge H, Keller K, Matzke W, Henning U, Schumann W (2004) Effect of dehulling of rapeseed on feed value and nutrient digestibility of rape products in pigs. Arch Anim Nutr 58:389–404
Liu L, Stein A, Wittkop B, Sarvari P, Li J, Yan X, Dreyer F, Frauen M, Friedt W, Snowdon R (2012) A knockout mutation in the lignin biosynthesis gene CCR1 explains a major QTL for acid detergent lignin content in Brassica napus seeds. Theor Appl Genet 124:1573–1586
Liu J, Osbourn A, Ma P (2015) MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol Plant 8:689–708
Liu X, Lu Y, Yan M, Sun D, Hu X, Liu S, Chen S, Guan C, Liu Z (2016) Genome-wide identification, localization, and expression analysis of proanthocyanidin-associated genes in Brassica. Front Plant Sci 7:1831
Meena PD, Awasthi RP, Chattopadhyay C, Kolte SJ, Kumar A (2016) Alternaria blight: a chronic disease in rapeseed-mustard. J. Oilseed Brassica 1:1–11
Meier U (2001) Entwicklungsstadien mono-und dikotyler Pflanzen. Die erweiterte BBCH monographie, 2. Gemeinschaftsarbeit der biologischen bundesanstalt für land- und forstwirtschaft (BBA), des bundessortenamtes (BSA) und des industrieverbandes agrar (IVA) unter Mitwirkung anderer Institutionen
Miao L, Chao H, Chen L, Wang H, Zhao W, Li B, Zhang L, Li H, Wang B, Li M (2019) Stable and novel QTL identification and new insights into the genetic networks affecting seed fiber traits in Brassica napus. Theor Appl Genet 132:1761–1775
Nagel M, Rosenhauer M, Willner E, Snowdon RJ, Friedt W, Börner A (2011) Seed longevity in oilseed rape (Brassica napus L.) – genetic variation and QTL mapping. Plant Genet Resour 9:260–263. https://doi.org/10.1017/S1479262111000372
Nagel M, Holstein K, Willner E, Börner A (2018) Machine learning links seed composition, glucosinolates and viability of oilseed rape after 31 years of long-term storage. Seed Sci Res 28:340–348
Qu C, Fu F, Lu K, Zhang K, Wang R, Xu X et al (2013) Differential accumulation of phenolic compounds and expression of related genes in black- and yellow-seeded Brassica napus. J Exp Bot 64:2885–2898. https://doi.org/10.1093/jxb/ert148
Qu C, Yin N, Chen S, Wang S, Chen X, Zhao H, Li J (2020) Comparative analysis of the metabolic profiles of yellow-versus black-seeded rapeseed using UPLC–HESI–MS/MS and transcriptome analysis. J agric food chem 68(10):3033–3049
Rahman MH (2001) Production of yellow-seeded Brassica napus through interspecific crosses. Plant Breed 120:463–472
Rahman M, McVetty P (2011) A review of Brassica seed color. Can J Plant Sci 91:437–446. https://doi.org/10.4141/CJPS10124
Rajjou L, Debeaujon I (2008) Seed longevity: survival and maintenance of high germination ability of dry seeds. CR Biol 331:796–805
Rashid A, Rakow G, Downey RK (1994) Development of yellow seeded Brassica napus through interspecific crosses. Plant Breed 112:127–134
Schatzki J, Allam M, Klöppel C, Nagel M, Börner A, Möllers C (2013) Genetic variation for secondary seed dormancy and seed longevity in a set of black-seeded European winter oilseed rape cultivars. Plant Breed 132:174–179
Simbaya J, Slominski BA, Rakow G, Campbell LD, Downey RK, Bello JM (1995) Quality characteristics of yellow-seeded Brassica seed meals: protein, carbohydrates, and dietary fiber components. J Agric Food Chem 43:2062–2066
Soomro TA, Ismail M, Anwar SA, Memon RM, Nizamani ZA (2020) Effect of Alternaria sp on seed germination in rapeseed, and its control with seed treatment. J Cereals Oilseeds 11:1–6
Stein A, Wittkop B, Liu L, Obermeier C, Friedt W, Snowdon RJ (2013) Dissection of a major QTL for seed colour and fibre content in Brassica napus reveals colocalization with candidate genes for phenylpropanoid biosynthesis and flavonoid deposition. Plant Breed 132:382–389
Stringam GR, McGregor DI, Pawlowski SH (1974) Chemical and morphological characteristics associated with seed-coat color in rapeseed. In: Proceeding of the 4th international rapeseed conference, Gießen, Germany. pp. 99–108
Sun F, Fan G, Hu Q, Zhou Y, Guan M, Tong C et al (2017) The high-quality genome of Brassica napus cultivar ‘ZS11’reveals the introgression history in semi-winter morphotype. Plant J 92:452–468
Tan SH, Mailer RJ, Blanchard CL, Agboola SO (2011) Canola proteins for human consumption: extraction, profile, and functional properties. J Food Sci 76:R16–R28
Taylor-Teeples M, Lin L, De Lucas M, Turco G, Toal TW, Gaudinier A et al (2015) An Arabidopsis gene regulatory network for secondary cell wall synthesis. Nature 517:571–575
Utz HF (2011) PLABSTAT: a computer program for statistical analysis of plant breeding experiments. Version 3A of 2011–06–14. http://www.uni-hohenheim.de/ipspwww/soft.html
Van Soest PJ, Robertson JB, Lewis BA (1991) Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci 74:3583–3597. https://doi.org/10.3168/jds.S0022-0302(91)78551-2
Wang J, Jian H, Wei L, Qu C, Xu X, Lu K, Qian W, Li J, Li M, Liu L (2015) Genome-wide analysis of seed acid detergent lignin (ADL) and hull content in rapeseed (Brassica napus L). PLoS ONE 10(12):e0145045. https://doi.org/10.1371/journal.pone.0145045
Wang J, Xian X, Xu X, Qu C, Lu K, Li J, Liu L (2017) Genome-wide association mapping of seed coat color in Brassica napus. J Agric Food Chem 65:5229–5237. https://doi.org/10.1021/acs.jafc.7b01226
Wei L, Jian H, Lu K, Yin N, Wang J, Duan X, Li W, Liu L, Xu X, Wang R, Paterson AH (2017) Genetic and transcriptomic analyses of lignin-and lodging-related traits in Brassica napus. Theor Appl Genet 130:1961–1973
Wittkop B, Snowdon RJ, Friedt W (2012) New NIRS calibrations for fiber fractions reveal broad genetic variation in Brassica napus seed quality. J Agric Food Chem 60:2248–2256
Yan XY, Li JN, Fu FY et al (2009) Co-location of seed oil content, seed hull content and seed coat color QTL in three different environments in Brassica napus L. Euphytica 170:355–364. https://doi.org/10.1007/s10681-009-0006-5
Zhang XK, Yang GT, Chen L, Yin JM, Tang ZL, Li JN (2006) Physiological differences between yellow-seeded and black-seeded rapeseed (Brassica napus L.) with different testa characteristics during artificial ageing. Seed Sci Technol 34:373–381
Zhou W, Chen F, Luo X, Dai Y, Yang Y, Zheng C, Yang W, Shu K (2020) A matter of life and death: Molecular, physiological, and environmental regulation of seed longevity. Plant Cell Environ 43:293–302
Acknowledgements
Andreas Gertz (KWS) and Frank Breuer (KWS) for performing field experiments and Illumina SNP chip analysis, Rosemarie Clemens for developing the DH populations, Sabine Freitag, Sibylle Pistrick (IPK), Gabriele Matzig (IPK) for excellent technical assistance, Laima Kott for providing seeds of DH1372, Boas Pucker for providing the annotation of the Brassica gene sequences, and Ilka Abreu for generating the PCA. IF acknowledges funding through the German Research Foundation (DFG: ZUK 45/2010). SW acknowledges the funding of RISET-PRO Ph.D scholarship program from Indonesian Ministry of Research and Technology.
Funding
Open Access funding enabled and organized by Projekt DEAL. IF received funding through the German Research Foundation (DFG: ZUK 45/2010). SW was supported by a RISET-PRO Ph.D scholarship program from Indonesian Ministry of Research and Technology.
Author information
Authors and Affiliations
Contributions
CM designed the project; SW performed the field experiments and did the NIRS analysis; SW, MN and AB did the seed germination tests, SW and CM analyzed the data, KF and IF performed and evaluated the non-targeted metabolite analysis; CM and KF wrote the manuscript; all authors have read and agreed on the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10681_2021_2891_MOESM1_ESM.xlsx
Data matrix of 1069 metabolite features (false discovery rate (FDR) < 0.005) obtained by metabolite fingerprinting (LC-HR-MS analysis) for genotype 1372 and DH-lines (4042 × Express 617) with either high or low ADLm content (ADL_High, ADL_Low) (XLSX 198 kb)
Variance components and heritabilities for seed quality traits of the two DH populations (PDF 33 kb)
10681_2021_2891_MOESM4_ESM.xlsx
Bulk specific SNP markers for the low and the high ADLm groups as detected following Illumina SNP chip analysis in the two DH populations (XLSX 14 kb)
10681_2021_2891_MOESM5_ESM.pdf
Metabolite fingerprinting of green seeds of the genotype DH1372 and DH-lines (4042 x Express 617) with either high or low ADLm content (ADL_High, ADL_Low) (PDF 159 kb)
10681_2021_2891_MOESM6_ESM.pdf
Depiction of the Principal component analysis (PCA) based on metabolite fingerprinting data of the genotype DH1372 and DH-lines (4042 x Express 617) with either high or low ADL content (ADL_High, ADL_Low) (PDF 128 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Widiarsih, S., Nagel, M., Börner, A. et al. Inheritance of seed quality and seed germination in two doubled haploid populations of oilseed rape segregating for acid detergent lignin (ADL) content. Euphytica 217, 161 (2021). https://doi.org/10.1007/s10681-021-02891-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-021-02891-z