Abstract
Bermudagrass is the most important warm-season pasture in the Southern USA with exceptional forage production potential and abiotic stress tolerance. However, it requires high nitrogen (N) supply to reach its full biomass and quality potential. Our objectives were to: (1) develop a nitrogen use efficiency (NUE) screening protocol for bermudagrass in controlled or semi-controlled conditions, (2) identify contrasting genotypes for NUE from natural variants and, (3) develop a knowledge base of NUE in bermudagrass. A collection consisting 290 Cynodon sp. genotypes was first pre-screened in the greenhouse. Thirty-nine genotypes with high NUE, five with low NUE were selected for further evaluations along with 5 checks in greenhouse and hoop-house under four N rates. Biomass, crude protein and N content were evaluated. N uptake efficiency (NUpE), N utilization efficiency (NUtE) and NUE were calculated based on biomass production. Genotypes showed significant influences (P < 0.0001) in all of the response variables. The genotype × N rate interaction was not significant for NUE in both environments. NUE had strong correlation with biomass production and NUpE, which got stronger as N rates increased. In N limiting conditions, bermudagrass showed a trade-off between biomass maintenance and crude protein content. Lower N applications increased biomass production over crude protein. However, when N is abundant the crop has the ability to improve crude protein. Several genotypes presented high NUE due their high NUtE and NUpE. Genotypes with contrasting NUE were selected and subjected to further field evaluation. Superior genotypes for NUE will be used in the breeding program to enhance NUE in bermudagrass.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Warm season perennial grasses are the foundation of pasture systems in the Southeast USA (Evers 1985) where cattle production is one of the main agricultural products (U.S. Census of Agriculture 2012). Among the tropical species, bermudagrass became the most important forage due its ability to adapt to a wide range of soils, aggressive growth, grazing tolerance, and ability to produce high yields of good quality forage when soil nutrients are not restricted (Redfearn and Rice 2014). The hybrid bermudagrass varieties e.g., Coastal, started to be developed in the 1940s (Burton 1948), which produced more than twice as much forage as common bermudagrass at moderate fertility levels (Burton and DeVane 1952). Since then several varieties have been selected for a broad range of desirable characteristics such as forage yield, crude protein, digestibility, winter hardiness, and disease resistance (Burton and Monson 1984, 1988; Burton et al. 1993; Taliaferro et al. 2002). A useful description of most bermudagrass forage cultivars is available at Corriher and Redmon (2011).
The forage varieties of bermudagrass are very responsive to nitrogen (N) fertilization (Burton and Jackson 1962). Fisher and Caldwell (1959) obtained maximum hay and protein production in an irrigated pasture when 1320 and 1490 kg ha−1 of N were applied, respectively. Prine and Burton (1956) showed that Coastal responded favorably to N fertilization up to 1009 kg ha−1 in a growing season with 990 mm of rainfall, but it reached less than half of that production in a dry year. Usually, N recommendations depend on the desired forage production. In general, it is recommended to apply at least 220 kg ha−1 of N since the availability of this nutrient is considered the most limiting factor for bermudagrass production (Redfearn et al. 2010).
Currently, less than 50% of the reactive N worldwide added onto cropland is converted into harvested products (Tilman et al. 2002; Fageria and Baligar 2005; Lassaletta et al. 2014). Pan et al. (2016) found that, globally, up to 64% (an average of 18%) of applied N was lost via NH3 volatilization, and the losses significantly increased with higher N application rate. Another part of the N applied is lost by Nitrate leaching, which is also a major worldwide cause of groundwater pollution (Sanz-Cobena et al. 2012). A crop that demands large amounts of N fertilization to reach full production implies large agronomic, economic, and energetic inefficiencies, as well as a large potential for excess N to be lost from crop fields causing environmental pollution (Nesheim et al. 2015).
The implementation of agricultural systems or crops with higher nitrogen use efficiency (NUE) is critical to reduce costs and maintain agricultural production in a sustainable way. The concept of NUE utilized in our work is the same described by Xu et al. (2012) which consists of the combination of nitrogen uptake efficiency (NUpE) and nitrogen utilization efficiency (NUtE). This process is governed by the interaction of multiple genetic and environmental factors (Xu et al. 2012). NUpE is the plant’s ability to absorb the N available in the soil. It is a function of root biomass, morphology, ability to explore regions with abundant nutrients and physiological capacity for nutrient uptake (Glass 2003). It also requires adequate sinks for temporary and long term nutrient storage (Hawkesford 2011). According to Dawson et al. (2008) increasing the recovery of N from fertilizer is an immediate way of improving agricultural NUE in crops. N losses by leaching and denitrification can be decreased by breeding crop varieties more efficient at capturing soil N during the entire growing season (Xu et al. 2012). NUtE, on the other hand, consists of the yield of the crop per unit of N acquired by the plant (Hawkesford 2011). When plants are growing in limited N conditions NUtE is an essential process and determines most of the variation in the NUE (Ågren 1985; Hirel et al. 2001).
N uptake and utilization efficiency are related to different morphological and physiological characteristics and are regulated by different genetic factors (Hirel et al. 2001; Masclaux-Daubresse et al. 2010), which enables the combination of favorable alleles when breeding for NUE. Increased N uptake and utilization efficiency may allow growers to maximize yield under a moderate rate of N fertilization instead of the traditional high rate of applications (Anbessa and Juskiw 2012).
The definition of NUE is not fixed and usually diverges from an ecological to agricultural point of view. Environmental conditions and characteristics of the crop also affect the importance of the different aspects of NUE. Therefore, the first step in a breeding project is to determine the main traits which can contribute to improvements in NUE and define the desired phenotype (Dawson et al. 2008). The current study is part of a bigger project which aims to select bermudagrass genotypes with higher NUE than the existing varieties based on the biomass production, yet, with minimal reduction in forage quality.
This paper investigates NUE in a collection of bermudagrass accessions. The objectives were to develop a NUE screening protocol for bermudagrass in controlled or semi-controlled conditions, identify contrasting genotypes for NUE from natural variants and, develop a knowledge base of NUE in bermudagrass.
Materials and methods
Selection of plant materials
The initial germplasm collection consisted of 290 genotypes of Cynodon dactylon L. and related species from 34 countries including di-, tri-, tetra-, penta- and hexa-ploid genotypes. Among these, 146 plant introductions (PIs) were from the bermudagrass core collection maintained at Tifton, GA, 137 PIs from the National Plant Germplasm System (NPGS) maintained at Griffin, GA and seven are commercial cultivars. A preliminary experiment was carried out in greenhouse to standardize a screening protocol and also to narrow down the number of genotypes. Based on this preliminary experiment data, 39 genotypes with high NUE, five genotypes with low NUE and five commercial cultivars were selected among the initial germplasm. The 49 genotypes selected based on NUE were utilized for further studies performed in greenhouse and hoop-house under four N rates, which are presented in this manuscript.
The whole germplasm collection (290 genotypes) has been maintained as clonal ramets in the Noble Research Institute greenhouse and is also being evaluated in an association mapping population (AMP) field study for NUE in bermudagrass.
Greenhouse and hoop house evaluations
For the greenhouse experiment, four bermudagrass ramets of each genotype were planted in cones (175 × 51 mm) filled up with 580 metro Mix. After 6 weeks of growth, the plants were transferred to 2.5 L vases (165 × 150 mm) containing 2.0 kg of pasteurized soil. The soil was collected from an Ardmore, OK field with inherently low in N (2 mg NO3 kg−1), 5.7 pH and 1.1% of organic matter. The soil was pasteurized to eliminate the presence of microorganisms and weed seed. The vases were placed in plastic saucers (140 × 80 mm) containing 3 cm of perlite. The vases fitted tightly in the saucers restricting evaporation and the perlite helped the roots to grow and reach extra solution leached from the vases. Top of vases were filled with 2 cm layer of perlite to reduce evaporation loss and moss growth. After transplanting, the plants were cut back and a nutrient solution (without N) was added to each pot depending on the initial soil sample analysis. The experiment was conducted following a split-plot complete randomized design (CRD) with 3 replications. N rates were considered as main plot and genotypes as sub-plot treatments. The greenhouse experiment set-up can be visualized in Fig. 1. The growth conditions were set at 32.2 °C day and 21.1 °C night temperature and a 16 h light regimen. The four N rates, as urea, were 0 (N0), 100 (N1), 200 (N2) and 400 (N3) mg N kg−1 soil (N+ treatments). Fertilizer was added in two split applications after first and second clippings. Biomass production was obtained by clipping the plants four times at 7 cm height with intervals of approx. 30 days. Fresh weights were taken and samples were dried at 75 °C for 72 h to obtain biomass production in grams of dry matter (DM). Only the above ground biomass, which is the plant part utilized as forage, was measured in our study. The total N content of the harvested samples was determined by high temperature combustion method (Leco Co. CNH-600 elemental analyzer, Saint Joseph, MI, at TAMU Soil, Water and Forage Testing Laboratory) and also estimated through NIRS (Near Infra-Red Spectroscopy). The crude protein content (%) was obtained multiplying the N content by 6.25.
An open-ended plastic-covered hoop-house located in the field was used to evaluate the plants (Fig. 1). The goal was to evaluate the plants in semi-field condition. The soil utilized in the hoop-house contained 0.5 mg NO3 kg−1, 7.7 pH and 1.7% of organic matter. Two 18.9 L buckets (368 × 260 mm) were utilized placing one bucket inside the other. The inside bucket contained six holes to allow the passage of solution and roots, and was filled with 18 kg of soil. The outside bucket (without holes) kept any solution leaching through the soil and made it available to the plant roots. One rooted ramet of each genotype was planted in cones (175 × 51 mm) containing 580 MetroMix. In genotypes with very small tillers, 2 tillers were used. Eight-week old plants were transferred to the buckets in hoop-house on April 28th, 2016. The experiment was done following split-plot design as discussed earlier. Weeds were manually removed each week. Watering was done manually and optimization of watering was done visually. The four N rates used in the hoop-house, as urea, were 0 (N0), 75 (N1), 150 (N2) and 300 (N3) mg N kg−1 soil (N+ treatments), which was divided in three equal applications. The N rates utilized in the greenhouse and hoop-house corresponded to very low (N0), insufficient (N1), sufficient (N2) and excessive (N3) N availability. In the hoop-house the plants were clipped on June 6th, June 29th, July 26th and August 25th. The data for biomass production and N content was obtained following the method described in the greenhouse experiment.
Calculating NUE
The NUE of the genotypes was calculated according to Moll et al. (1982), biomass produced per unit of N available in the soil, with minor modifications. NUE is the product of NUpE and NUtE. Where, NUpE is the capacity of plant roots to acquire N from the soil. And, NUtE is the plant biomass productivity per unit of N uptake (Nup), calculated following the protocol of Hawkesford (2011). The Nup is N contained in the plant biomass. The amount of N available (Nav) in the soil is quantified as the sum of the N from fertilizer applied (Nf) plus the N uptake by aboveground plant tissues (Nt) in pots with no N fertilizer applied (N0) (Fageria and Baligar 2005).
The formulas used are:
Statistical analysis
The data sets from hoop-house and greenhouse were analyzed separately. The response variables, biomass and crude protein, were analyzed combining N0 and N+ traits (all N rates) and also only with N+ rates in order to better visualize the effect of N fertilization and its interactions. The response variables NUE, NUpE and NUtE were calculated for N+ rates. A split-plot on completely randomized design (CRD) was employed to investigate the effects of N rates and genotypes on each response variable. In the statistical design, N rates and genotypes were applied to main-plot and sub-plot, respectively. The replicates were treated as random factor. The analyses were performed with proc glm procedure (SAS 9.3). Significance was declared if P < 0.05. The minimum LSD statistics was approximately calculated for each trait since all sample sizes are not the same for all genotypes. The R2 of the model was calculated for each response variable. Pearson correlations among response variables were performed using SAS 9.3.
Results and discussion
Greenhouse and hoop-house evaluations
Highly significant differences (P <0.0001) were obtained among the genotypes for all response variables in the greenhouse. The N rates also had significant effects on the majority of the variables (Table 1). The exception was for NUpE (P =0.1848, Table 1) with no differences among the N rates. In the greenhouse, interactions between genotypes and N rates were significant for biomass (all N rates), crude protein (all N rates and N+) and NUtE (Table 1). In the hoop-house, highly significant differences (P <0.001) were obtained between genotypes and N rates for all variables evaluated (Table 1). More significant genotype × N rate interactions were obtained in the hoop-house. These were significant for biomass (all N rates and N+), crude protein (all N rates and N+), NUpE and NUtE (Table 1).
No significant genotype × N rate interaction was observed for NUE in either greenhouse (P = 0.24) or hoop-house (P = 0.41) indicating that N use efficient genotypes are the same in low and high N rates. These results are different from that obtained in rice (Oriza sativa L.) (Singh et al. 1998; Wu and Tao 1995) and maize (Zea mays L.) (Gallais and Hirel 2004; Coque and Gallais 2007), where significant genotype × N rates interactions for NUE have been found. However, despite the significant interaction, Singh et al. (1998) observed that six out of twenty rice genotypes had high NUE in low N input as well as in high N input. In wheat, varieties that responded well in low N-inputs also performed well in high N inputs (Ortiz-Monasterio et al. 1997). In forages, studies comparing NUE among genotypes are scarce. In a related approach to NUE, Wilkins et al. (2000) evaluated eight perennial ryegrass varieties at five levels of N fertilizer. Rank of the varieties was similar for all five N levels for annual biomass yield and N content.
Biomass and crude protein (at all N rates and N+ rates), NUtE and NUE presented high R2 values (R2 > 80%) in both environments for the statistical model utilized (Table 2), which indicates that most of the variation was explained by the model. NUpE had R2 close to 80% in hoop-house (R2 = 79) as well, however the value for NUpE in greenhouse was the lowest (R2 = 66%) among all studied variables (Table 2). NUpE is directly related with the root’s ability to uptake nutrient and depends on root biomass, morphology and physiology (Glass 2003). The lower R2 value, no-significant differences among N rates and, no-significant N rate x genotype interaction for NUpE only in greenhouse could be an effect of the small containers utilized in this environment (2.5 L vases). The smaller container possibly restricted the root development especially for the genotypes with higher root growth potential. Poorter et al. (2012) stated the importance of appropriate pot size since higher plant mass per pot volume reduces plant growth and can affect differences between treatments. In hoop-house, we used 18.9 L vases thus seemed to have less limitations for shoot and root development offering more adequate conditions for NUE studies in bermudagrass.
Biomass production
As expected, the N fertilization strongly affects biomass production with significant increments as more N was applied (Table 1, Fig. 2a, b). On average, the highest relative increments in biomass were obtained at N1 compared to the plants at N0 (95% in hoop-house and 174% in greenhouse) (Fig. 2a, b, Online resource 2 and 3). The relative increments tended to be smaller as higher N rates were applied (13% in the hoop-house and 21% in greenhouse from N2 to N3) tending to stabilization. In greenhouse the genotype 272 had the highest biomass production differing from Tifton-85 which was the best control across all N rates (Fig. 2a, Table 3). On average, high NUE genotypes produced more biomass in all N conditions, especially at N0 in greenhouse and N3 in hoop-house (Fig. 2a, b). The highest biomass was obtained by different genotypes in each N rate in the hoop-house (116 at N0 and N2, 241 at N1 and 275 at N3). Among the controls, Jiggs had the highest biomass in hoop-house followed by B2000 and Tifton-85 (Fig. 2b, Online resource 3). The cultivar Tifton-85, broadly used for forage, presented high biomass production (26% more than Coastal) and high digestibility (11% more than Coastal) when evaluated in a 3-year trial in Georgia, USA (Burton et al. 1993). Jiggs can produce equal amounts of biomass to Tifton-85 having advantage on sites that are tight and poorly drained (Corriher and Redmon 2011).
Biomass (g) (a, b) and crude protein (%) (c, d) of 49 bermudagrass genotypes evaluated in greenhouse (left panel) and hoop-house (right panel) under four different N rates. Green and orange points correspond to the five genotypes with the highest and lowest NUE, respectively, in each environment. Blue points corresponds to the controls. Grey points are all the other genotypes. The line graphs show the trends of the highest NUE genotypes (272 in greenhouse and 275 in hoop-house) and lowest (264 in greenhouse and 55 in hoop-house) and the controls (Tifton-85 and Jiggs). The color of the line for the highest and lowest NUE genotypes varies according to the ranking of the genotype in each environment (green, orange or grey)
Crude protein content
The addition of N also significantly increased the protein content of bermudagrass in all N rates (Table 1, Fig. 2c, d). The complete data set of mean values for crude protein of the genotypes within N rates can be visualized in the Online resources 4 and 5. Overall, the genotypes with high NUE presented the lowest protein in N0, N1 and N2 (Fig. 2c, d). However, these genotypes improved their performance in relation to the other genotypes at N3 (Fig. 2c, d). The genotype 272, for example, increased protein content only by 1% at N1 compared to N0, staying under 10% of protein in both environments. However, the increments went up to 3% at N2 and 5% at N3 (Fig. 2c, d). At the highest N rate, genotype 272 had protein content higher than 17% close to the average of all genotypes in both environments. The genotype with the highest NUE in hoop-house (275) also improved in protein from N0 (8.9%) to N3 (16%). However, 275 did not increase its protein as much as 272 did, having the third lowest protein overall (Fig. 2d). Neither check cultivars nor high NUE genotypes seemed to be close to a plateau for protein at N3, indicating potential to even higher increments. Most of the low NUE genotypes, even though they had better protein in low N rates, did not have high potential to improve the crude protein with the addition of N fertilizer (Fig. 2c, d).
While for biomass the highest increments occurred at N1 in relation to N0, for crude protein the N1 rate provided the lowest increments especially for high NUE genotypes (Fig. 2). Fisher and Caldwell (1959) also obtained more significant increments in biomass production than protein when low N rates were applied in a bermudagrass pasture. Burton and DeVane (1952) reported low increments in protein content in split applications of 56 and 112 kg ha−1 of N while heavy applications of N (448 kg ha−1) significantly increased protein. These results indicate that in N limiting conditions bermudagrass seems to prioritize biomass production over crude protein. However, when more N is available bermudagrass has the ability to improve crude protein content. In our study, we also observed that some of the high NUE genotypes have higher potential to improve crude protein than others with similar NUE. The selection of genotypes with this potential would enable to obtain genotypes with superior NUE without losing forage quality.
N uptake efficiency
In the greenhouse there was high variability among the genotypes in all N rates for NUpE (Fig. 3a). On average the highest NUpE in greenhouse was 75% at N2 dropping to 66% at N3 as more N was available with no statistical significance (Table 1, Online resource 6). However, the genotype 272, Jiggs and Tifton-85 maintained NUpE between 75 and 80% across all the N rates (Fig. 3a). Even though there were significant differences among the genotypes for NUpE in the greenhouse the majority of the genotypes belonged to a common group, especially at N1 and N2 with LSD of 19% (Fig. 3a, Table 3). Among the controls, Midland had lower NUpE (65.2%) than the others at N1. Tifton-85, Jiggs, B2000 and Cheyenne did not differentiate from the first ranked genotypes in all N rates (307 at N1, 334 at N2 and 277 at N3).
Nitrogen uptake (a, b), nitrogen utilization (c, d) and nitrogen use (e, f) efficiencies of the 49 bermudagrass genotypes evaluated in greenhouse (left panel) and hoop-house (right panel) under three different N rates. Green and orange points correspond to the five genotypes with the highest and lowest NUE each, respectively, in greenhouse and hoop-house. Blue points corresponds to the controls. Grey points are all the other genotypes. The line graphs show the trends of in the highest (272 in greenhouse and 275 in hoop-house) and lowest (264 in greenhouse and 55 in hoop-house) NUE genotypes and the controls (Tifton-85 and Jiggs). The color of the line for the highest and lowest NUE genotypes varies according to the ranking of the genotype in each environment (green, orange or grey)
Increased N availability significantly reduced the NUpE across N rates in hoop-house (Table 2, Fig. 3b). In the N1 treatment, on average 87% of the available N was uptaken by the plants. At high N rate (N3), the NUpE dropped to 61%. In hoop-house, variation for NUpE among all genotypes was smaller at low N rates, which indicated that most of the available N was up taken by the genotypes (Fig. 3b). As more N was added, the variability increased and only a few genotypes had the ability to achieve a NUpE above 75% (Fig. 3b, Online resource 7). In Arabidopsis it was also observed that the variation in NUE at high N inputs was mostly due to variation in NUpE (Chardon et al. 2010). At N3 all low NUE genotypes had NUpE below 55% while the high NUE genotypes maintained NUpE between 70 and 80% (Fig. 3b). Tifton-85 had a slight decrease in NUpE from N1 (84.3%) to N2 (81.4%), however at N3, the NUpE drastically decreased to 57% (Fig. 3b). The genotype 275, with the highest NUE in hoop-house, was stable in the two lowest N levels (89%) and decreased only to 76.5% at the highest N rate. Brentrup and Pallière (2010) affirms that a scenario of very high as well as very low NUpE values represent unsustainable crop production. NUpE values between 90 and 100% represent a risk of soil mining, where the N required by the plant is not met by the N input. NUpE values below 70% bring an increased risk of N losses and should be avoided to protect the environment (Brentrup and Pallière 2010). A well-balanced N input and output would have NUpE of 80–90%, which was obtained, in our hoop-house study at N2 (150 mg N kg−1 soil) by most of the genotypes with high NUE. However, the most sustainable N rate for each genotype can only be determined evaluating NUpE in field conditions.
N utilization efficiency
The NUtE showed an opposite pattern of variability compared to that observed in NUpE, especially in the hoop-house (Fig. 3d). At the lowest N rate the genotypes differed by more than 20 g in NUtE. However, the variability among genotypes decreased to less than 10 g in N3 (Fig. 3c, d). These results agree with Coque and Gallais (2007), who stated that at low N-inputs, both components (NUpE and NUtE) are involved in the NUE, however NUtE had the main role. In our study, the high NUE genotypes presented NUtE values above average especially in N1 treatment in both the experiments (Fig. 2c, d). These results indicated that high NUE genotypes could produce double the biomass per unit of N uptake than low NUE genotypes especially in N limiting conditions. The NUtE values at each N rate were very similar between the greenhouse and hoop-house evaluations, showing consistency of the genotypes independent of the environments (Fig. 3c, d, Online resources 8 and 9). The highest NUtE obtained was 67.1 (g g−1) at N1, by 272 in the greenhouse and 223 in the hoop-house. The main attributes that will provide higher NUtE are photosynthetic activity, canopy size, longevity, and sink organ capacity (Hawkesford 2011).
NUtE is the biomass productivity per unit of N content of the above ground plant parts (Hawkesford 2011). Thus, inevitably the higher the NUtE, the lower the N content of the plant, in other words, the forage nutritive values. Sinebo et al. (2004) observed in barley that genotypes with the highest grain yield had the lowest N concentration, with N concentration negatively correlated to NUE. The improvement of NUE should not be achieved at the expense of other agronomic traits such as forage quality. Therefore, it is essential to better understand the physiological basis of the relationship between NUE, biomass, and N concentration under limiting and non-limiting N conditions to achieve a successful selection (Bélanger and Gastal 2000).
N use efficiency
NUE of the genotypes drastically decreased as more N was added to the plants (Fig. 3e, f, Table 1). On average, plants grown in a low N rate produced double biomass per unit of Nav compared to the highest N rate. Tilman et al. (2002) reviewed that in general, the highest NUE occurs at lowest N rate and decreases as more N is added. Lower NUE at higher N rates can indicate that plants cannot absorb and/or utilize all the N added or that the N losses exceeded the rate of plant uptake (Fageria and Baligar 2005).
Variability for NUE within species was reported in several crops: spring barley (Hordeum vulgare L.) (Anbessa et al. (2009), rice (Singh et al. 1998; Wu and Tao 1995), winter wheat (Triticum aestivum L.) (Van Sanford and MacKown 1986), maize (Coque and Gallais 2007). In our study significant differences between genotypes were also observed for NUE (Table 1). In greenhouse the genotype 272 obtained the highest NUE values in all N rates with 55.1, 43.4 and 28 (g g−1) at N1, N2 and N3, respectively (Fig. 3e, f). On average, 272 was 10% more N use efficient than the next ranked genotype (genotype 8) in all N rates, however the difference was not statistically significant (P < 0.05) (Fig. 3e, Table 3). When compared to the check cultivars, 272 did not differentiate from Tifton-85 and Jiggs in any N rates (Fig. 3e, Table 3) and from B2000 at N3. Midland and Cheyenne had significantly lower NUE in all N rates (Online resource 10). In the hoop-house genotype 275 ranked on the top with 56.5, 42.7, 29.1 (g g−1) at N1, N2 and N3, respectively. On average 275 was 6.1% and 9.6% more efficient than the cultivars Jiggs and Tifton-85, respectively, without statistical difference (Fig. 3f, Table 3). The check cultivars Tifton-85, Jiggs and B2000 consistently showed high NUE in both greenhouse and hoop-house experiments while Midland and Cheyenne had significantly lower NUE (Online resource 10 and 11). A separation from the high and low NUE genotypes across all N rates can be visualized in the Fig. 3e, f, especially in hoop-house, which reflects the absence of interaction between N rate and genotypes. Genotypes with higher NUE at both low as well in high N rates are the most desirable in a breeding program (Fageria and Baligar 2005). Data averaged across greenhouse and hoop-house experiments showed that the genotypes 272, 8, 241, 322, 223, 102 were ranked among the top-15 in both environments. Tifton-85 ranked 3rd in the greenhouse and 12th in the hoop-house, while Jiggs ranked 9th in both experiments. Genotypes with the highest NUE across all N rates and also those showing the highest NUE in different environments can be advantageous from a breeding perspective and can be utilized as parents in future breeding programs.
The mean values for NUE and NUpE found in this study were notably higher than those normally obtained in other crops. It has been documented that in cereal crops, i.e., wheat, corn, rice, barley, sorghum (Sorghum bicolor L.), millet (Pennisetum glaucum (L.) oat (Avena sativa L.) and, rye (Secale cereale L.), less than 50% of the N fertilizer applied is transformed into harvestable products (Raun and Johnson 1999; Tilman et al. 2002; Fageria and Baligar 2005; Lassaletta et al. 2014). However, higher NUE has been obtained in cereal species when utilized as forage than for grain, like in wheat–rye–ryegrass pasture (Altom et al. 1996) and corn silage (O’Leary and Rehm 1990). Thomason et al. (2000) observed that wheat utilized for forage had higher total N uptake and NUE than wheat cultivated for grain production in all four N rates evaluated. The higher N uptake in forages may be a consequence of the harvests before flowering, when N losses are greater (Altom et al. 1996; Hooker et al. 1980). Another reason for higher NUE in forages can be as a result of the shoots being the final harvested product and they include the majority of the plant N (Gargallo-Garriga et al. 2014). In grain crops, besides the NUpE and NUtE, N remobilization efficiency to the grains is a crucial factor for NUE (Masclaux-Daubresse et al. 2010; Avice and Etienne 2014). In grazing crops, any increase in NUpE or/and NUtE can directly increase NUE leading to a higher biomass production, better quality or both. Consequently, NUE in forages can be a simpler trait to be selected.
Relationship among N efficiencies
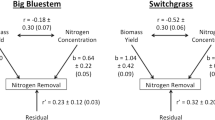
The Pearson correlation analysis indicated that most of the correlations among biomass, NUtE, NUpE, NUE and NUp are significant and positive in both environments (Table 4, Online resource 12). The exceptions are correlations between NUtE versus NUpE and NUtE versus NUp. Crude protein is negatively correlated with NUtE, biomass and NUE (Table 4). Although, NUE is negatively correlated with crude protein in both experiments, these negative correlations are at least 20% lower in N3 compared to N1. NUE has high positive correlation with biomass in all N+ treatments, and the relationship became stronger as higher amounts N were added and reached 99% at the N3 in both experiments. NUE also had a high positive correlation with NUpE, which followed the similar pattern as with biomass and the correlation increases from N1 to N3 in greenhouse (83–90%) and hoop-house (70–94%). In contrast, NUE × NUtE correlations tend to decrease as more N was applied. These results showed advantage in selecting genotypes in higher N rates. Since NUE does not have significant genotype × N rate interaction an efficient selection for high NUE plants could be done independently of the N rate. Besides that, a selection in higher N rates would increase the probability to select the genotypes with highest biomass, NUpE and with higher potential to increase crude protein content when N is not limited. Selecting for biomass yield can effectively select for the combination of NUtE and NUpE which are very separate traits (Hawkesford 2011). According to Moll et al. (1982) it is important to find an optimum fertility environment or develop a selection index based on data at several fertility levels in order to ensure equal selection pressure on NUtE and NUpE to effectively select for NUE.
Bermudagrass has been considered a low NUE crop due the high amounts of N rates that are usually recommended. However, some genotypes with high NUE and NUpE were identified even at the highest N rates. Selection of genotypes with efficient N uptake and utilization efficiency is an important strategy, among many others, to achieve higher NUE and sustainability of the agricultural system (Fageria and Baligar 2005; Anbessa and Juskiw 2012). Large improvements in NUE are necessary to increase livestock and crop production in a sustainable way in order to meet future global needs (Tilman et al. 2002). In this study we observed high potential to optimize NUE in bermudagrass through selection and breeding of genotypes with the right traits. To continue this process genotypes with contrasting NUE were selected based on the results presented here and are being evaluated in field conditions for NUE.
Conclusions
We developed a protocol that enabled the screening and selection of bermudagrass genotypes for contrasting NUE. The bigger vases seemed to have less limitations for shoot and root development offering more adequate conditions for NUE studies in bermudagrass. There was no significant genotype × N rate interaction for NUE in bermudagrass under greenhouse and hoop-house conditions. The bermudagrass germplasm presented high variability for NUE. Some genotypes have high NUE due their high NUtE, as well as high NUpE even at the highest N rate. Genotypes with high NUE tend to have the lowest crude protein content especially in the low N rates. However, some of these genotypes might have a superior potential to improve crude protein in non-restrictive N conditions than others.
References
Ågren GI (1985) Theory for growth of plants derived from the nitrogen productivity concept. Physiol Plant 64(1):17–28
Altom W, Rogers JL, Ram WR, Johnson GV, Taylor SL (1996) Long-term rye–wheat–ryegrass forage yields as affected by rate and date of applied nitrogen. J Prod Agric 9(4):510–516
Anbessa Y, Juskiw P (2012) Strategies to increase nitrogen use efficiency of spring barley. Can J Plant Sci 92(4):617–625
Anbessa Y, Juskiw P, Good A, Nyachiro J, Helm J (2009) Genetic variability in nitrogen use efficiency of spring barley. Crop Sci 49(4):1259–1269
Avice J-C, Etienne P (2014) Leaf senescence and nitrogen remobilization efficiency in oilseed rape (Brassica napus L.). J Exp Bot 65(14):3813–3824
Bélanger G, Gastal F (2000) Nitrogen utilization by forage grasses. Can J Plant Sci 80(1):11–20
Brentrup F, Pallière C (2010) Nitrogen use efficiency as an agro-environmental indicator. In: Proceedings of the OECD workshop on agrienvironmental indicators, March, 2010, pp 23–26
Burton GW (1948) Coastal bermuda grass. Ga. Coastal Plain Expt. Sta. Cir., vol 10
Burton GW, DeVane EH (1952) Effect of rate and method of applying different sources of nitrogen upon the yield and chemical composition of bermudagrass, Cynodon dactylon (L) Pers., and hay. Agron J 44:5
Burton GW, Jackson JE (1962) Effect of rate and frequency of applying six nitrogen sources on coastal bermudagrass. Agron J 54(1):40–43
Burton GW, Monson WG (1984) Registration of Tifton 68 bermudagrass. Crop Sci 24:1211
Burton GW, Monson WG (1988) Registration of ‘Tifton 78’ bermudagrass. Crop Sci 28(1):187–188
Burton GW, Gates RN, Hill GM (1993) Registration of ‘Tifton 85’ bermudagrass. Crop Sci 33(3):2
Chardon F, Barthélémy J, Daniel-Vedele F, Masclaux-Daubresse C (2010) Natural variation of nitrate uptake and nitrogen use efficiency in Arabidopsis thaliana cultivated with limiting and ample nitrogen supply. J Exp Bot 61(9):2293–2302
Coque M, Gallais A (2007) Genetic variation among European maize varieties for nitrogen use efficiency under low and high nitrogen fertilization. Maydica 52(4):383
Corriher VA, Redmon LA (2011) Bermudagrass varieties, hybrids and blends for Texas. Tec. Rep E-320 9(11)
Dawson JC, Huggins DR, Jones SS (2008) Characterizing nitrogen use efficiency in natural and agricultural ecosystems to improve the performance of cereal crops in low-input and organic agricultural systems. Field Crops Res 107(2):89–101
Evers G (1985) Forage and nitrogen contributions of arrowleaf and subterranean clovers overseeded on bermudagrass and bahiagrass. Agron J 77(6):960–963
Fageria N, Baligar V (2005) Enhancing nitrogen use efficiency in crop plants. Adv Agron 88:97–185
Fisher F, Caldwell A (1959) The effects of continued use of heavy rates of fertilizers on forage production and quality of coastal bermudagrass. Agron J 51(2):99–102
Gallais A, Hirel B (2004) An approach to the genetics of nitrogen use efficiency in maize. J Exp Bot 55(396):295–306
Gargallo-Garriga A, Sardans J, Pérez-Trujillo M, Rivas-Ubach A, Oravec M, Vecerova K, Urban O, Jentsch A, Kreyling J, Beierkuhnlein C (2014) Opposite metabolic responses of shoots and roots to drought. Sci Rep 4:6829
Glass AD (2003) Nitrogen use efficiency of crop plants: physiological constraints upon nitrogen absorption. Crit Rev Plant Sci 22(5):453–470
Hawkesford MJ (2011) An overview of nutrient use efficiency and strategies for crop improvement. In: Hawkesford MJ, Barraclough P (eds) The molecular and physiological basis of nutrient use efficiency in crops. Wiley-Blackwell, West Susses, pp 3–19
Hirel B, Bertin P, Quilleré I, Bourdoncle W, Attagnant C, Dellay C, Gouy A, Cadiou S, Retailliau C, Falque M (2001) Towards a better understanding of the genetic and physiological basis for nitrogen use efficiency in maize. Plant Physiol 125(3):1258–1270
Hooker M, Sander D, Peterson G, Daigger L (1980) Gaseous N losses from winter wheat. Agron J 72(5):789–792
Lassaletta L, Billen G, Grizzetti B, Anglade J, Garnier J (2014) 50 year trends in nitrogen use efficiency of world cropping systems: the relationship between yield and nitrogen input to cropland. Environ Res Lett 9(10):105011
Masclaux-Daubresse C, Daniel-Vedele F, Dechorgnat J, Chardon F, Gaufichon L, Suzuki A (2010) Nitrogen uptake, assimilation and remobilization in plants: challenges for sustainable and productive agriculture. Ann Bot 105(7):1141–1157
Moll R, Kamprath E, Jackson W (1982) Analysis and interpretation of factors which contribute to efficiency of nitrogen utilization. Agron J 74(3):562–564
Nesheim MC, Oria M, Yih PT (eds) (2015) Committee on a framework for assessing the health, environmental, and social effects of the food system. Tood and Nutrition Board, Board on Agriculture and Natural Resources, Institute of Medicine, National Research Council, Bethesda
O’Leary M, Rehm G (1990) Nitrogen and sulfur effects on the yield and quality of corn grown for grain and silage. J Prod Agric 3(1):135–140
Ortiz-Monasterio R, Sayre K, Rajaram S, McMahon M (1997) Genetic progress in wheat yield and nitrogen use efficiency under four nitrogen rates. Crop Sci 37(3):898–904
Pan B, Lam SK, Mosier A, Luo Y, Chen D (2016) Ammonia volatilization from synthetic fertilizers and its mitigation strategies: a global synthesis. Agr Ecosyst Environ 232:283–289
Poorter H, Bühler J, van Dusschoten D, Climent J, Postma JA (2012) Pot size matters: a meta-analysis of the effects of rooting volume on plant growth. Funct Plant Biol 39(11):839–850
Prine GM, Burton GW (1956) The effect of nitrogen rate and clipping frequency upon the yield, protein content and certain morphological characteristics of Coastal bermudagrass (Cynodon dactylon, (L) Pers.). Agron J 48(7):296–301
Raun WR, Johnson GV (1999) Improving nitrogen use efficiency for cereal production. Agron J 91(3):357–363
Redfearn D, Rice C (2014) Bermudagrass pasture management PSS-2591. Accessed 25th Sept 2017
Redfearn D, Arnall B, Zhang H, Rice C (2010) Fertilizing bermudagrass hay and pasture. PSS-2263 Oklahoma Coop Ext, Oklahoma State Univ, Stillwater
Sanz-Cobena A, Sánchez-Martín L, García-Torres L, Vallejo A (2012) Gaseous emissions of N2O and NO and NO3 − leaching from urea applied with urease and nitrification inhibitors to a maize (Zea mays) crop. Agr Ecosyst Environ 149:64–73
Sinebo W, Gretzmacher R, Edelbauer A (2004) Genotypic variation for nitrogen use efficiency in Ethiopian barley. Field Crops Res 85(1):43–60
Singh U, Ladha J, Castillo E, Punzalan G, Tirol-Padre A, Duqueza M (1998) Genotypic variation in nitrogen use efficiency in medium- and long-duration rice. Field Crops Res 58(1):35–53
Taliaferro C, Anderson J, Richardson W, Baker J, Coleman S, Phillips W, Sandage L, Moyer J, Hansen T, Kallenbach R (2002) Registration of Midland 99′ forage bermudagrass (Registrations Of Cultivars). Crop Sci 42(6):2212–2214
Thomason W, Raun W, Johnson G (2000) Winter wheat fertilizer nitrogen use efficiency in grain and forage production systems. J Plant Nutr 23(10):1505–1516
Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S (2002) Agricultural sustainability and intensive production practices. Nature 418(6898):671–677
U.S. Census of Agriculture (2012) United States Department of Agriculture, Washington, DC
Van Sanford D, MacKown C (1986) Variation in nitrogen use efficiency among soft red winter wheat genotypes. Theor Appl Genet 72(2):158–163
Wilkins P, Allen D, Mytton L (2000) Differences in the nitrogen use efficiency of perennial ryegrass varieties under simulated rotational grazing and their effects on nitrogen recovery and herbage nitrogen content. Grass Forage Sci 55(1):69–76
Wu P, Tao Q (1995) Genotypic response and selection pressure on nitrogen-use efficiency in rice under different nitrogen regimes. J Plant Nutr 18(3):487–500
Xu G, Fan X, Miller AJ (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63:153–182
Acknowledgments
The authors are very thankful to James Rogers and his group, Noble Research Institute for assistance in hoop-house experiment. We are grateful to Forage Analysis and Analytical Core facilities of the Noble Research Institute for sample analysis. Funding for this project was provided through Forage365 Initiative of the Noble Research Institute, 2510 Sam Noble Parkway, Ardmore, OK 73401, USA.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Schneider-Canny, R., Chekhovskiy, K., Muñoz, P. et al. Characterization of bermudagrass (Cynodon dactylon L.) germplasm for nitrogen use efficiency. Euphytica 215, 40 (2019). https://doi.org/10.1007/s10681-019-2347-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-019-2347-z