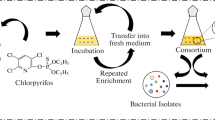
Abstract
Pollutants cause a huge problem for humans, animals, plants, and various ecosystems, especially water resources. Agricultural, domestic, and industrial waste effluents change the water quality and affect living microorganisms. Therefore, the current study aimed to identify possible microorganisms in wastewater as potential bioremediation agents of pesticide residues. Wastewater samples were collected from El-Khairy agricultural drainage, which receives agricultural and domestic wastes. Bacteria and fungi species were isolated as clean cultures. Wastewater samples were analyzed for pesticide residues via gas chromatography-mass spectroscopy (GC–MS) system. Results uncovered the presence of ten pesticides ranging from 0.0817 to 28.162 µg/l, and the predominant pesticide was chlorpyrifos. Along with that, about nine species (3 bacterial and 6 fungal) were relatively efficient in the removal of chlorpyrifos residues up to 2000 µg/l with removal percentages ranging from 24.16 to 80.93% under laboratory conditions. Two bacterial isolates proficiently degraded significant amounts of chlorpyrifos: Bacillus cereus strain PC2 (GenBank accession No. MZ314010) and Streptomyces praecox strain SP1 (GenBank accession No. MZ314009). In-site bacterial and fungal isolates defined in the current study were proficient in cleaning wastewater of chlorpyrifos pesticide residues.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Water shortage and quality deterioration, along with the ever-increasing population, would be great challenges facing many countries worldwide (Abdel-Gawadh et al., 2004). By 2050, about 6 billion people will suffer from the scarcity of clean water (Boretti & Rosa, 2019; UN-WWDR, 2020). In Egypt, water shortage is an alarming problem; its effects have been growing in recent years. Egypt depends mainly on the Nile River to sustain freshwater supplies (Dakkak, 2020). Since 1959, the Nile provides Egypt with about 55.5 billion m3/year; hence, the water share per capita has been reduced from 2560 m3/year in 1959 to 980 m3/year in 2000 and is expected to reach 637 m3/year by 2025 (Ashour et al., 2003).
Along with that, water resources are polluted with industrial, domestic, and agricultural waste (Ashour et al., 2003; Koshal, 1976). For example, heavy metals, asbestos, nitrates, detergents, solvents, fertilizers, and pesticides were listed as major pollutants of the River Nile (El-Sheekh, 2009). Pesticides cause numerous negative health and environmental effects (Nicolopoulou-Stamati et al., 2016). Especially, the long-lasting pesticides in ecosystems, for instance, organochlorine insecticides were found in water resources after more than 20 years of their use (Caughey, 1999; Seo et al., 2007). Specifically, organophosphorus pesticides contaminated various ecosystems around the world and caused adverse effects to millions of people with over 200,000 deaths annually (Abraham et al., 2013).
Furthermore, improper disposal and overuse of pesticides have added large amounts to water and soil environments. Pesticides might reach water through runoff with irrigation water, air drifting, leaching, and/or direct application (dusting and spraying), which in turn affect the water quality and aquatic organisms (Abbassy et al., 2020). Consequently, human exposure to these pesticides’ residues might occur throughout food chain leading to various adverse impacts (Agrawal et al., 2010; Hakeem et al., 2016). Therefore, cleaning water resources through the implementation of proper remediation methods is extremely needed. Specifically, the strategy of biological remediation (bioremediation) is widely employed. Because it depends mainly on the nature and type of pollutants and the metabolic degradation mechanism of microbes (Megharaj et al., 2011; Moss, 2008). Also, its applications are rapidly adapted as a suitable alternative to conventional clean-up technologies (Vidali, 2001), where microorganisms are more adjustable to environmental changes and deterioration (Vroumsia et al., 2005).
Moreover, the bioremediation rate and level of a pesticide depend on its bioavailability, uptake rate by the microbiological cell, and the growth rate of the cells with the pesticide as the energy source (Abatenh et al., 2017; Odukkathil & Vasudevan, 2013). Recently, diverse organisms, including algae, bacteria, and fungi and plants were employed to clean polluted environments of pesticides (Díaz, 2004; Pushpanathan et al., 2014; Vay et al., 2001). Specifically, several bacterial strains showed a degradation potential of organophosphate insecticides (Cycoń et al., 2009). Pseudomonas fluorescens, Brucella melitensis, Bacillus subtilis, and P. aeruginosa were able to remove 89, 87, 85, and 92% of chlorpyrifos (CPF), respectively, after 30 days of incubation (Lakshmi et al., 2008). Also, Bacillus sp., Brevundimonas sp., Pseudomonas sp., Sphingomonas sp., and Stenotrophomonas sp. significantly degraded from 37 to 100 mg/l/d of CPF (Li et al., 2008). The P. aeruginosa, B. cereus, Serratia marcescens, and Klebsiella sp. were effective in removing 84, 84, 81, and 80%, respectively, of CPF from liquid media after 20 days, while after 30 days in soils, they removed about 92, 60, 56, and 37%, respectively (Lakshmi et al., 2009). Similarly, Lactobacillus lactis, L. fermentum, and E. coli efficiently converted CPF to its oxon and DETP metabolites (Harishankar et al., 2013).
The utilization of pesticides by microorganisms as sources of minerals (carbon and phosphorous) and energy would help in cleaning various water resources from their residues. Therefore, the current study aimed to isolate indigenous bacteria and fungi and screen their potential as bioremediation agents of wastewater of pesticide residues.
Materials and methods
Chemicals
Acetonitrile HPLC-grade and culture media were purchased from local chemical providers. DNA purification kit (Germany) and PCR clean-up kit were from Maxim Biotech Inc. (USA). The internal standard (TPP) and extraction (Cat#5982–0650) and dispersive SPE clean-up (Cat#5982–5056) kits were purchased from Technoscient for Lab & Optical Product, Cairo, Egypt. Certified reference standard materials of pesticides were obtained from ULTRA Scientific Analytical Solutions (RI, USA) (Table 1).
Source of water samples
Wastewater samples were collected from three locations: start, middle, and end at the El-Khairy drainage, El-Beheira Governorate, north of Egypt (Fig. 1). The drainage receives industrial, domestic, and agricultural effluents. Its water is being re-used for the irrigation of vast areas of agricultural lands; the total served area is 27,500 feddan. It is considered the main agricultural drainage of a length of 21.65 km and receives an average discharge of 14.2 m3/s (Veeningen, 1999).
Sampling of wastewater
Subsurface water samples (25 cm below the surface) were collected by water sampler into cleaned and sterilized one-liter Pyrex borosilicate dark glass bottles (Radwan et al., 2019). Water bottles were used for the bacteria and fungi screening and isolation and pesticide residue analyses. Samples were transferred to the Pesticides Residues Analysis and Toxicity Laboratory (PRATL), Faculty of Agriculture, Damanhour University, for analysis within a few hours of collection.
Isolation of microorganisms from wastewater
Microorganisms found in wastewater samples were separated and sub-cultured on potato dextrose agar for fungi isolation (200 g of infusion from potatoes, 20 g of dextrose, 15 g of agar at pH 5.6 ± 0.2) and plate count agar for isolation of the bacteria colonies (5 g of tryptone, 2.5 g of yeast extract, 1 g glucose, and 15 g agar at pH 7.0 ± 0.25) at 25 °C. Nine samples were collected; 3 from each collection point as replicates and 3 plates per replicate were planted from each sample under sterilized conditions and incubated at 37 °C. The complete growth of the microbe was reached after about 7 days of incubation. Each microorganism was transferred into a new Petri dish. The subculture of each microbe was repeated several times until a visually clean culture was obtained.
Analysis of pesticide residues in wastewater using GC–MS
Pesticide extraction and clean-up
Pesticide residues in wastewater were extracted and cleaned up using a modified method of Anastassiades (Anastassiades et al., 2003). Extraction (Cat#5982–0650) and dispersive SPE clean-up (Cat#5982–5056) kits of Agilent Technologies were used. Specifically, about 10 ml of wastewater was vortexed with 10 ml of 0.1% acidified MeCN for 1 min. Then 4 g and 1 g of MgSO4 (anhy) and NaCl, respectively, were thoroughly mixed for 1 min. Then the internal standard triphenyl phosphate (TPP) solution was added to tubes and shaken for 30 s. Then tubes were centrifuged at 1350 × g for 10 min (Hermle Labortechnik GmbH, Siemensstr 25 D-78564 Wehingen, Germany). About 1 ml of supernatant (acetonitrile) was mixed by hand for 5 min with 25 mg PSA sorbent and 150 mg MgSO4 (anhy) and centrifuged for 5 min at 1350 × g. About 500 µl of each tube was filtered through 0.22-μm PTFE filters (Millipore, USA) into HPLC vials for GC–MS analysis.
Separation and determination conditions
Separation and identification of residues in wastewater samples and recoveries were accomplished using an Agilent GC–MS system in split-less mode. Exactly, 2 µl of each sample was injected into an HP-5MS capillary column (30 m × 0.53 mm i.d. 0.25 µm film thickness). Separation conditions were as reported by AOAC (AOAC, 2007), where the initial temperature of the column was set at 80 °C for 6 min, increased at 15 °C/min to 215 °C (held for 1 min), and then the column was heated to 230 °C at 5 °C/min and to 290 °C at 5 °C/min (held for 2 min). The carrier gas was the helium gas at a constant flow rate of 1.1 ml/min. The observed pesticides were identified by the full mass spectra scans using the total ion chromatogram (TIC) and search of spectra in the EI-MS libraries. Concentrations of identified pesticides were calculated based on a standard curve of each compound.
Quality control parameters
The intra-day assay (repeatability) and inter-day assay (intermediate precision) of the used analytical technique were calculated according to Ermer (2005). Also, the precision expressed as coefficients of variation, limits of detection (LOD), and quantification (LOQs) were estimated. For recovery studies, water samples were fortified with 0.1 and 1 µg/l of each standard and analyzed following the above-mentioned methods. Then percentages of recovery samples were reported ± relative standard deviation (%RSD) (Frenich et al., 2006). All detected amounts of pesticides residues were corrected considering the recovery percentages (Urovic & Orevi, 2011).
Microorganisms’ tolerance assay
The radial growth of separated and identified fungi and bacteria on media mixed with the prevalent insecticide in wastewater, chlorpyrifos (CPF), was examined. Five concentrations of CPF (0, 100, 500, 1000, and 2000 µg/l) were prepared in the growing media. Five plates of each microorganism were used as replicates per each concentration. Microorganisms were incubated with the CPF at 37 °C for 1 week, and then their radial growth was photographed and recorded. Then competence of growth fungi and bacteria on such media with the insecticide was calculated compared to control plates. This experiment was repeated six times. Then the performance of the potential bioremediation activity of organisms was examined using 500, 1000, and 2000 µg/l. The residues of CPF in media after the incubation time were measured using the GC–MS as described previously.
Identification of fungal isolates
The fungal isolates were cultured onto clean growth media until pure cultures were obtained and used for various evaluations and identification (Lichtwardt, 1985). Microscopic observations were done on mounted cultures using lactic acid. The morphological traits of each fungal colony were observed and recorded as described in fungal atlases (Klich, 1990).
Identification of bacterial isolates
Taxonomic characterization of isolated bacteria was conducted at the genus level based on morphological, physiological, and biochemical traits and following the method reported in the manuals of Bergey of Systematic Bacteriology (James, 1998) and Cowan and Steel’s (Barrow & Feltham, 1993).
Molecular characterization of bacterial isolates
Extraction of genomic DNA
Two bacterial isolates were chosen to be identified genetically because of their elevated potential in the degradation of CPF insecticide. The DNA of isolated bacteria was extracted using Qiagen DNA kit (Qiagen, Hilden, Germany) following the manufacturer’s guidelines.
Amplification of the 16S rRNA
About 350 bp of 16 s rRNA gene was amplified. The 350F and 350R primers that correspond to the 16S rRNA conserved gene sequence of E. coli, forward, 5ʹ AGG ACG TGC TCC AAC CGC A ʹ3, and reverse, 5ʹ AAC TGG AGG AAG GTG GGG AT ʹ3 (Sambrook & Russell, 2001). The PCR reaction was as the following: an initial cycle of 95 °C for 5 min and 34 cycles of 95 °C for 1 min, 47 °C for 1 min, and 72 °C for 1 min and then an extension cycle at 72 °C for 10 min. Then amplified products were visualized on 1% agarose gel, stained by ethidium bromide, and photographed using a gel documentation system. Then the purification of amplified PCR products was done using PCR clean-up column kit (Maxim Biotech INC, USA).
Sequencing and alignment
The 16S gene DNA sequence (excised and purified bands) was performed by Macrogen Company (Seoul, South Korea). The 16S rRNA gene nucleotide sequences of isolated bacterial strains were submitted to the GenBank database under accession numbers MZ314009 and MZ314010.
Sequence alignment and phylogenetic analysis
Pair-wise and multiple DNA sequence alignments were performed using CLUSTALW program version 1.82 (http://www.ebi.ac.uk/clustalw) (Thompson et al., 1994). Neighbor-joining trees were created using MEGA version 6 (Tamura et al., 2013) from the CLUSTALW for each strain. Comparison between obtained DNA sequence alignments and sequences in GenBank database was completed using BLASTN searches at http://ncbi.nlm.nih.gov.
Statistical analysis
Experimental data were statistically analyzed using the SAS software (SAS, Cary, USA, version 9.3). The pesticide residues and microbial isolates’ performance and growth were expressed as mean ± SD. Significant means were contrasted using Tukey’s honest significant difference test (HSD) (P ≤ 0.05) (SAS, 2013).
Results
Pesticide residues in wastewater
Quality control limits
The analytical method used in the analysis of pesticides was accurate and suitable based on obtained values of repeatability and intermediate precision (Ermer, 2005). Results of the intra- and inter-assay ranged from 3.47–6.18% and 6.51–10.11%, respectively, for detected pesticides (Table 2). These results were within the acceptable range set by residue analysis laboratories. Also, recovery results of detected pesticides at 0.5 and 5 µg/l levels ranged from 87.65–93.57% and 91.73–95.37%, respectively.
Pesticide residues in wastewater samples
Analysis of pesticides residues revealed the presence of lenacil, chlorpyrifos, cypermethrin, bifenthrin, carbofuran, and permethrin at 0.721, 28.16, 4.14, 0.052, 7.881, and 0.208 µg/l, respectively, in samples collected from site A (Table 2). Chlorpyrifos, cypermethrin, bifenthrin, tolfenpyrad, and permethrin were found in samples from site B with concentrations of 16.05, 3.24, 0.09, 1.02, and 0.09 µg/l, respectively. Samples from site C had oxamyl, dicloran, simetryn, sulfotep, chlorpyrifos, cypermethrin, and ethofumesate at 5.071, 0.0817, 2.3045, 1.0251, 23.087, 2.3408, and 0.0981 µg/l, respectively. Chlorpyrifos insecticide was the dominant compound in the wastewater samples. Therefore, it was selected for the bioremediation examinations.
Microorganisms and chlorpyrifos removal
The results of the growth of separated fungi and bacteria on media mixed with the prevalent insecticide in wastewater (CPF) were presented in Table 3. Five concentrations of CPF (0, 100, 500, 1000, and 2000 µg/l) were added to the growing media. Five Petri dishes of each microorganism were used as replicates per each concentration. The growth of isolated organisms was recorded (Table 3) and photographed (Fig. 2) after 1 week of incubation with or without the insecticide. The microorganisms A, D, and F showed the best growth on plates with or without CPF. The F bacteria was the most grown on media with 2000 µg/l followed by A and D up to 1000 µg/l of CPF compared to all isolated bacteria and fungi (Fig. 2 and Table 3).
Verification of the performance of isolated bacteria (D, F, and I) and fungi (A, B, E, G, H, and J) was examined by challenging their growth on media with 500, 1000, and 2000 µg/l of CPF (Fig. 3). Results showed good potential to use these species in the degradation of CPF up to 1000 µg/l with efficiency (% of removal) ranging from 31.41 (for H) to 90.82% (for F). Specifically, F, I, and A removed about 80.82, 80.93, and 75.59% of the 2000 µg/l of CPF after 1 week.
Percentages of removal of chlorpyrifos insecticide by isolated species after 7 days of incubation at 37° C. N = 3 Petri plates per replicate, 4 replicates per concentration. Isolated organisms code: A: Aspergillus terreus, B: Aspergillus foetidus var. pallidus, D: Bacillus cereus, E: Aspergillus fumigatus var. ellipticus, F: Streptomyces praecox, G: Aspergillus fumigates, H: Penicillium janthinellum, I: Unidentified bacteria, J: Aspergillus fumigatus var. ellipticus
Identification of fungal isolates
Six different fungal species were identified according to the growth and microscopic characteristics (Table 4 and Fig. 4). Identification was done based on guidelines of the Regional Center for Mycology and Biotechnology (RCMB) using image analysis protocols. The fungal isolates A, B, E and J, and G were Aspergillus terreus, Aspergillus foetidus var. pallidus, Aspergillus fumigatus var. ellipticus, and Aspergillus fumigates according to the database identification program of RCMB For Aspergilli (Klich, 1990). The sixth fungal isolate (H) was Penicillium janthinellum; identification was conducted based on current universal keys as described in fungal atlases (James, 1998).
Identification of bacterial isolates
The characteristics of the most efficient bacterial isolates (F and I) in degrading CPF were listed in Table 5. The motility, gram staining, sporulation, growth at 40 °C, anaerobic growth, Kovac’s oxidase, gelatin liquefaction, starch hydrolysis, and acid production from lactose, mannose, arabinose, maltose, and sucrose were listed. The isolates were identified as Streptomyces praecox (F) and Bacillus cereus (I). For further differentiation between the two bacterial strains, similar morphology, physiological, and biochemical traits were reported, except for starch hydrolysis and lactose production that were negative in B. cereus and positive in S. praecox. Also, B. cereus produced acid from sucrose, but S. praecox did not.
Phylogenetic identification of bacteria
The 16S rRNA gene sequences of Bacillus cereus PC2 (GenBank Acc# MZ314010) and Streptomyces praecox SP1 (GenBank Acc# MZ314009) were constructed via molecular identification. Results of DNA sequences of the 16S gene of F and I bacterial isolates were as the following for F, identified as Streptomyces praecox, with a sequence of the following:
-
GACGGCCTTCGGGTTGTAAACCTCGGGCAGCAGGGAAGAAGCGCAAGTGACGGTACCTGCAGAAGAAGCG
-
CCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGCCCCAAGCGTTGTCCGGAATTATTGGGCGTA
-
AAGAGCTCGTAGGCGGCTTGTCACGTCGGATGTGAAAGCCCGGGGCTTAAGGGGGGGTCTGCATTCGATA
-
CGGGCTAGCTAGAGTGTGGTAGCCCAGATCGGAATTCCTGGTGTAGCGGTGAAATGCGCAGATATCAGGA
-
GGAACACCGGTGGCGTTGGCGGATCTCTGGGCCATTACTGACGCTGAGGAGCGAAAGCGTGGGGAGCGAA
-
CAGGATTAGATACCCTGGTAGTCCACGCCGTAAACGTTGGGAACTAGGTGTTGGCGACATTCCACGTCGT
-
CGGTGCCGCAGCTAACGCATTAAGTTGGGGGCCTGGGGAGTACGGCCGCAAGGCTAAAACTCAAAGGAAT
The second bacteria (I) was identified as Bacillus cereus with a DNA sequence of the following:
-
CAGACTCCTACGGGAGGCAGCAGTAGGGAATCTTCCGCAATGGACGAAAGTCTGACGGAGCAACGCCGCG
-
TGAGTGATGAAGGCTTTCGGGTCGTAAAACTCTGTTGTTAGGGAAGAACAAGTGCTAGTTGAATAAGCTG
-
GCACCTTGACGGTACCTAACCAGAAAGCCACGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGTG
-
GCAAGCGTTATCCGGAATTATTGGGCGTAAAGCGCGCGCAGGTGGTTTCTTAAGTCTGATGTGAAAGCCC
-
ACGGCTCAACCGTGGAGGGTCATTGGAAACTGGGAGACTTGAGTGCAGAAGAGGAAAGTGGAATTCCATG
-
TGTAGCGGTGAAATGCGTAGAGATATGGAGGAACACCAGTGGCGAAGGCGACTTTCTGGTCTGTAACTGA
-
CACTGAGGCGCGAAAGCGTGGGGAGCAAACAGGATTAGATACCCTGGTAGTCCACGCCGTAAACGATGAG
-
TGCTAAGTGTTAGAGGGTTTCCGCCCTTTAGTGCTGAAGTTAACGCATTAAGCACTCCGCCTGGGGAGTA
The identified 16S rRNA gene sequence of S. praecox strain SP1 was compared with the other 16 species of Streptomyces in GenBank. The neighbor-joining phylogenetic tree revealed high homology between S. praecox strain SP1 and S. praecox strain 7445 (Fig. 5). Also, the phylogenetic tree of the gene sequence of Bacillus cereus strain PC2 with the other 25 strains of B. cereus strains in the GenBank revealed high homology with B. cereus strain IAM 12,605 (Fig. 6).
Phylogenetic tree showing the evolutionary relationship between Streptomyces praecox 16S rRNA nucleotide sequence genes (Strain SP1; accession number MZ314009) and the other Streptomyces sp. presented in the GenBank. The tree dendrogram was constructed using the neighbor-joining method using the Mega software version 6
Phylogenetic tree showing the evolutionary relationship between Bacillus cereus 16S rRNA nucleotide sequence genes (strain PC2; accession number MZ314010) and the other B. cereus presented in the GenBank. The tree dendrogram was constructed using the neighbor-joining method using the Mega software version 6
Discussions
Indigenous microorganisms in water might define the fate of applied pesticides, undergoing degradation, transport, and adsorption/desorption processes (Kaur & Garg, 2014). The intact and degraded products of pesticides affect, significantly, these microorganisms, thus altering their microbial diversity and might cause pollution (Díaz-Cruz & Barceló, 2006). Chlorpyrifos was first introduced in 1965 by Dow Chemicals in the USA to manage a diverse number of agricultural and domestic insects (John & Shaike, 2015). It is extensively used in Egypt to control various insects and mites on a variety of field crops and residential plants (Aly et al., 2021). But it might adversely disturb ecosystems through serious environmental pollution (Cycoń et al., 2009; Liu et al., 2012). CPF was reported to cause adverse effects to pesticide applicators and farmers during its application (Farahat et al., 2010). Furthermore, pollution with CPF might affect microorganisms and non-target bees, wasps, and aquatic organisms (Jabeen et al., 2015), where it persists in neutral soil with a half-life ranging from 35 to 78 days (at 25 °C). But when it was used as a termiticide, CPF remains for 175–1576 days (Solomon et al., 2014).
The biodegradation process of CPF intermediates with the formation of 3,5,6-trichloro-2-pyridinol (TCP) as the main metabolite with greater water solubility compared to TCP. Streptomyces sp. strain JAAS1 was effective in degrading both CPF and TCP in contaminated sites (Abraham et al., 2013). Also, there were numerous reports on the employment of microorganisms in the detoxification of CPF from water, for example, P. aeruginosa (NCIM 2074) (Fulekar & Geetha, 2008) and the G1 strain of Stenotrophomonas sp. efficiently degraded eight OPs due to their versatile systems of genes and enzymes (Deng et al., 2015). The B. cereus, B. subtilis, B. melitensis, Klebsiella sp., P. aeruginosa, P. fluorescence, and S. marcescens were used CPF as main source of carbon (Lakshmi et al., 2008). In another study, Sphingomonas sp., Stenotrophomonas sp., Bacillus sp., Brevundimonas sp., and Pseudomonas sp. were removed from 37 to 100 mg/l/day of CPF individually. Sphingomonas sp. showed the greatest activity in the transformation of CPF (100 mg/l) within 24 h (Li et al., 2008). Also, Stenotrophomonas sp. PF32 used about 97% of 100 mg/l of CPF in 48 h as a carbon source. Moreover, the S. maltophilia strain MHF ENV20 degraded half of the amounts of CPF and its metabolites, trichlorophenol and diethyl thiophosphate salt in 96 h (Deng et al., 2015).
The Actinobacteria, Streptomyces sp. AC5 and AC7 species, effectively biodegraded CPF insecticide. The AC5 removed over 90% of CPF (50 mg/l) after 72 h. of incubation, while the AC7 strain was less effective (Briceño et al., 2012). Similarly, Synechocystis sp. eliminated 3.78 and 4.69 mg/l of 5 mg/l of CPF within 4 and 8 days, respectively (Singh et al., 2011). Both Pennisetum pedicellatum and Stenotrophomonas maltophilia transformed CPF to TCP and DETP and utilized them as carbon, phosphorous, and nitrogen sources (Dubey & Fulekar, 2012). Along with those, several bacterial strains were effective in degrading CPF and used it as a source of carbon, for example, B. pumilus (Anwar et al., 2009), Flavobacterium sp. (Mallick et al., 1999), E. coli (Richins et al., 1997), Alcaligenes faecalis DSP3 (Yang et al., 2005), Klebsiella sp. (isolated from wastewater) (Ghanem et al., 2007), Providencia stuartii (Bhatia, 2008), P. aeruginosa, and Clavibacter michiganensis (Subhas & Singh, 2003).
Similar to the results reported herein, there are a few fungal strains that were capable of degrading CPF. For example, Verticillium sp. eliminated up to 90% of CPF within 7 days at pH 7 at 35 °C (Fang et al., 2008; Yu et al., 2006). The fungal species, Aspergillus sp., Penicillium sp., Eurotium sp., and Emericella sp., degraded about 70% of CPF within 1 week and used it as a carbon and nitrogen source (Xu et al., 2007). Cladosporium cladosporioides Hu-01 mineralized CPF after 6 days of incubation (Chen et al., 2012). Aspergillus terreus-JAS1 dissociated CPF and TCP in liquid media and soil after 24 h of incubation (Silambarasan & Abraham, 2013). Compared to bacterial isolates, fungal species efficiently bioremediate CPF via a mineralization mechanism (Supreeth & Raju, 2017).
Conclusion
Pollution of water resources with pesticides might cause serious problems. Indigenous microorganisms help lessen the adverse effects of pesticide residues through degradation. Agricultural wastewater samples were analyzed for pesticide residues, and results showed the detection of lenacil, chlorpyrifos, cypermethrin, bifenthrin, carbofuran, tolfenpyrad, oxamyl, dicloran, simetryn, sulfotep, ethofumesate, and permethrin. Chlorpyrifos insecticide was the dominant compound in wastewater samples. Indigenous bacterial and fungal species were isolated, and their ability to degrade chlorpyrifos insecticide was examined. After 1 week of incubation of isolated bacteria (D, F, and I) and fungi (A, B, E, G, H, and J) with CPF, results revealed efficiency % of removal ranging from 31.41 to 90.82%. Specifically, F (Streptomyces praecox), I (Bacillus cereus), and A (Aspergillus terreus) removed about 80.82, 80.93, and 75.59% of the 2000 µg/l of CPF in 1 week. Following the molecular identification of these species, they were registered in the GenBank as Streptomyces praecox strain SP1 and Bacillus cereus strain PC2 with accession Nos. MZ314009 and MZ314010, respectively.
Data availability
All data and materials have been presented in the manuscript.
References
Abatenh, E., Gizaw, B., Tsegaya, Z., & Wassie, M. (2017). Application of microorganisms in bioremediation-review. Journal of Environmental Microbiology, 1(1): 1–9
Abbassy, M., Khalifa, M., Nassar, A., Omar, A., Salim, Y., & Nour-Eldin, E. (2020). Comparative study of organochlorine pesticide residues in water of both Edko Lake and Fish Farm in El-Behira. Austin Environmental Sciences, 5(1), 1044.
Abdel-Gawadh, S. T., Kandil, M., & Sadek, T. M. (2004). Water scarcity prospects in Egypt 2000 – 2050. In Environmental Challenges in the Mediterranean 2000–2050. Nato Science Series, IV. Earth and Environmental Sciences Volume 37 https://doi.org/10.1007/978-94-007-0973-7_12
Abraham, J., Shanker, A., & Silambarasan, S. (2013). Role of Gordonia sp JAAS1 in biodegradation of chlorpyrifos and its hydrolysing metabolite 3,5,6-trichloro-2-pyridinol. Letters in Applied Microbiology. https://doi.org/10.1111/lam.12141
Agrawal, A., Pandey, R. S., & Sharma, B. (2010). Water pollution with special reference to pesticide contamination in India. Journal of Water Resource and Protection, 02(05), 432–448. https://doi.org/10.4236/jwarp.2010.25050
Anastassiades, M., Lehotay, S. J., & Stajnbaher, D. (2003). Quick, easy, cheap, effective, rugged, and safe (QuEChERS) approach for the determination of pesticide residues. 18th Annual Waste Testing and Quality Assurance Symposium, WTQA 2002 - Proceedings, 231–241.
Anwar, S., Liaquat, F., Khan, Q. M., Khalid, Z. M., & Iqbal, S. (2009). Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol by Bacillus pumilus strain C2A1. Journal of Hazardous Materials. 168(1): 400–405. https://doi.org/10.1016/j.jhazmat.2009.02.059
AOAC. (2007). AOAC Official Method 2007.01 pesticide residues in foods by acetonitrile extraction and partitioning with magnesium sulfate gas chromatography/mass spectrometry and liquid chromatography/tandem mass spectrometry first action 2007. AOAC International. https://www.academia.edu/33471645/AOAC_Official_Method_2007.01_Pesticide_Residues_in_Foods_by_Acetonitrile_Extraction_and_Partitioning_with_Magnesium_Sulfate_Gas_Chromatography_Mass_Spectrometry_and_Liquid_Chromatography_Tandem_Mass_Spectrometry_First_Act (Accessed June 2020)
Ashour, M. A., El Attar, S. T., Rafaat, Y. M., Mohamed, M. N., & Manger, T. M. (2003). Water resources management II. Progress in Water Resources, 8(2), 269–279.
Barrow, G., & Feltham, R. (1993). Cowan and steel’s manual for the identification of medical bacteria (3rd ed.). Cambridge University Press. https://doi.org/10.1136/jcp.46.10.975-a
Bhatia, D. (2008). Isolation and characterization of a chlorpyrifos-degrading bacterium from agricultural soil and its growth response. African Journal of Microbiology Research, 2(26), 31.
Boretti, A., & Rosa, L. (2019). Reassessing the projections of the World Water Development Report. npj Clean Water 2(15). https://doi.org/10.1038/s41545-019-0039-9
Briceño, G., Fuentes, M. S., Palma, G., Jorquera, M. A., Amoroso, M. J., & Diez, M. C. (2012). Chlorpyrifos biodegradation and 3,5,6-trichloro-2-pyridinol production by actinobacteria isolated from soil. International Biodeterioration and Biodegradation 73: 1–7. https://doi.org/10.1016/j.ibiod.2012.06.002
Caughey, M. E. (1999). Pesticides in surface waters: Distribution, trends, and governing factors; volume 3 of the series pesticides in the hydrologic system. Steven J. Larson, Paul D. Capel and Michael S. Majewski, Ann Arbor Press, Inc., Chelsea, MI, (1997), 390 Pages, [ISBN No. Environmental Progress. https://doi.org/10.1002/ep.670180107
Chen, S., Liu, C., Peng, C., Liu, H., Hu, M., & Zhong, G. (2012). Biodegradation of chlorpyrifos and its hydrolysis product 3,5,6-trichloro-2-pyridinol by a new fungal strain Cladosporium cladosporioides Hu-01. PLoS ONE (10): e47205.. https://doi.org/10.1371/journal.pone.0047205
Cycoń, M., Wójcik, M., & Piotrowska-Seget, Z. (2009). Biodegradation of the organophosphorus insecticide diazinon by Serratia sp. and Pseudomonas sp. and their use in bioremediation of contaminated soil. Chemosphere 76(4): 494–501. https://doi.org/10.1016/j.chemosphere.2009.03.023
Dakkak, A. (2020). Egypt’s water crisis – recipe for disaster. EcoMENA. https://www.ecomena.org/egypt-water/. Accessed Jun 2020.
Deng, S., Chen, Y., Wang, D., Shi, T., Wu, X., Ma, X., et al. (2015). Rapid biodegradation of organophosphorus pesticides by Stenotrophomonas sp: G1. Journal of Hazardous Materials, 297, 17–24. https://doi.org/10.1016/j.jhazmat.2015.04.052
Díaz-Cruz, M. S., & Barceló, D. (2006). Highly selective sample preparation and gas chromatographic-mass spectrometric analysis of chlorpyrifos, diazinon and their major metabolites in sludge and sludge-fertilized agricultural soils. Journal of Chromatography A 1132(1–2, 3): 21–27. https://doi.org/10.1016/j.chroma.2006.07.062
Díaz, E. (2004). Bacterial degradation of aromatic pollutants: A paradigm of metabolic versatility. International Microbiology 7(3):173–80. https://doi.org/10.2436/im.v7i3.9468
Dubey, K. K., & Fulekar, M. H. (2012). Chlorpyrifos bioremediation in Pennisetum rhizosphere by a novel potential degrader Stenotrophomonas maltophilia MHF ENV20. World Journal of Microbiology and Biotechnology 28:1715–1725. https://doi.org/10.1007/s11274-011-0982-1
El-Sheekh, M. (2009). River Nile pollutants and their effect on life forms and water quality. In The Nile: Origin, Environments, Limnology and Human Use (pp. 395–405). Springer Science + Business Media B.V. https://doi.org/10.1007/978-1-4020-9726-3_19
Ermer, J. (2005). Analytical validation within the pharmaceutical environment. In Method Validation in Pharmaceutical Analysis: A Guide to Best Practice.Eds: Ermer, J. & McB. Miller J.H. Print ISBN:9783527312559, Online ISBN:9783527604685. https://doi.org/10.1002/3527604685.ch1
Fang, H., Qin Xiang, Y., Jie Hao, Y., Qiang Chu, X., Dong Pan, X., Quan Yu, J., & Long Yu, Y. (2008). Fungal degradation of chlorpyrifos by Verticillium sp. DSP in pure cultures and its use in bioremediation of contaminated soil and pakchoi. International Biodeterioration and Biodegradation 61(4): 294–303. https://doi.org/10.1016/j.ibiod.2007.10.001
Farahat, F. M., Fenske, R. A., Olson, J. R., Galvin, K., Bonner, M. R., Rohlman, D. S., et al. (2010). Chlorpyrifos exposures in Egyptian cotton field workers. Neurotoxicology, 31(3), 297–304. https://doi.org/10.1016/j.neuro.2010.02.005
Frenich, A. G., Vidal, J. L. M., González, F. J. E., & Liébanas, F. J. A. (2006). Quality criteria in pesticide analysis. In Methods in Biotechnology, Pesticide Protocols (Vol. 19, pp. 219–230). https://doi.org/10.1385/1-59259-929-x:219
Fulekar, M. H., & Geetha, M. (2008). Bioremediation of chlorpyrifos by Pseudomonas aeruginosa using scale up technique. Applied Biosciences 12: 657–660.
Ghanem, I., Orfi, M., & Shamma, M. (2007). Biodegradation of chlorpyrifos by Klebsiella sp. isolated from an activated sludge sample of waste water treatment plant in Damascus. Folia Microbiologica 52: 423–427. https://doi.org/10.1007/BF02932098
Hakeem, K. R., Akhtar, M. S., & Abdullah, S. N. A. (2016). Plant, soil and microbes: Volume 1: Implications in crop science. Plant, Soil and Microbes: Volume 1: Implications in Crop Science, (April), 1–366. https://doi.org/10.1007/978-3-319-27455-3
Harishankar, M. K., Sasikala, C., & Ramya, M. (2013). Efficiency of the intestinal bacteria in the degradation of the toxic pesticide, chlorpyrifos. 3 Biotech. https://doi.org/10.1007/s13205-012-0078-0
Jabeen, H., Iqbal, S., & Anwar, S. (2015). Biodegradation of chlorpyrifos and 3, 5, 6-trichloro-2-pyridinol by a novel rhizobial strain Mesorhizobium sp. HN3. Water and Environment Journal 29(1): 151–160. https://doi.org/10.1111/wej.12081
James M. J. (1998). Modern Food Microbiology, 5th edition. A Chapman & Hall Food Science Book (Vol. 373). https://doi.org/10.1016/S0140-6736(09)60492-X
John, E. M., & Shaike, J. M. (2015). Chlorpyrifos: Pollution and remediation. Environmental Chemistry Letters 13: 269–291. https://doi.org/10.1007/s10311-015-0513-7
Kaur, H., & Garg, H. (2014). Pesticides: Environmental impacts and management strategies. Pesticides - Toxic Aspects. https://doi.org/10.5772/57399
Klich, M. A. (1990). Computer applications in Penicillium and Aspergillus systematics. In Modern Concepts in Penicillium and Aspergillus Classification (pp. 269–278). Springer US.https://doi.org/10.1007/978-1-4899-3579-3_23
Koshal, R. K. (1976). Water pollution and human health. Water, Air, and Soil Pollution, 5(3), 289–297. https://doi.org/10.1007/BF00158344
Lakshmi, C. V., Kumar, M., & Khanna, S. (2008). Biotransformation of chlorpyrifos and bioremediation of contaminated soil. International Biodeterioration and Biodegradation, 62(2), 204–209. https://doi.org/10.1016/j.ibiod.2007.12.005
Lakshmi, C. V., Kumar, M., & Khanna, S. (2009). Biodegradation of chlorpyrifos in soil by enriched cultures. Current Microbiology 58: 35–38. https://doi.org/10.1007/s00284-008-9262-1
Li, X., Jiang, J., Gu, L., Ali, S. W., He, J., & Li, S. (2008). Diversity of chlorpyrifos-degrading bacteria isolated from chlorpyrifos-contaminated samples. International Biodeterioration and Biodegradation 62(4):331–335. https://doi.org/10.1016/j.ibiod.2008.03.001
Lichtwardt, R. W. (1985). The Trichomycetes. Fungal Associates of Arthropods. Springer-Verlag. https://doi.org/10.1007/978-3-540-79307-6_1
Liu, Z., Chen, X., Shi, Y., & Su, Z. C. (2012). Bacterial degradation of chlorpyrifos by Bacillus cereus. Advanced Materials Research. Conference: International Conference on Energy, Environment and Sustainable Volume: 356–360. https://doi.org/10.4028/www.scientific.net/AMR.356-360.676
Mallick, K., Bharati, K., Banerji, A., Shakil, N. A., & Sethunathan, N. (1999). Bacterial degradation of chlorpyrifos in pure cultures and in soil. Bulletin of Environmental Contamination and Toxicology, 62(1), 48–54. https://doi.org/10.1007/s001289900840
Megharaj, M., Ramakrishnan, B., Venkateswarlu, K., Sethunathan, N., & Naidu, R. (2011). Bioremediation approaches for organic pollutants: A critical perspective. Environment International, 37(8), 1362–1375. https://doi.org/10.1016/j.envint.2011.06.003
Nicolopoulou-Stamati, P., Maipas, S., Kotampasi, C., Stamatis, P., & Hens, L. (2016). Chemical pesticides and human health: The urgent need for a new concept in agriculture. Frontiers in Public Health, 4(July), 1–8. https://doi.org/10.3389/fpubh.2016.00148
Odukkathil, G., & Vasudevan, N. (2013, December 20). Toxicity and bioremediation of pesticides in agricultural soil. Reviews in Environmental Science and Biotechnology. Springer. https://doi.org/10.1007/s11157-013-9320-4
Pushpanathan, M., Jayashree, S., Gunasekaran, P., & Rajendhran, J. (2014). Microbial bioremediation: A metagenomic approach. In Microbial Biodegradation and Bioremediation 407–419. https://doi.org/10.1016/B978-0-12-800021-2.00017-0
Radwan, E., Eissa, E., Nassar, A. M. K., Salim, Y., Hashem, H., Abdul-Aziz, K., & Abdel-Hakeem, N. (2019). Study of water pollutants in El-Mahmoudia agricultural irrigation stream at El-Beheira Governorate, Egypt. J Bioinform Syst Biol, 2(1), 1–018. https://doi.org/10.26502/fjbsb004
Richins, R. D., Kaneva, I., Mulchandani, A., & Chen, W. (1997). Biodegradation of organophosphorus pesticides by surface-expressed organophosphorus hydrolase. Nature Biotechnology 15: 984–987. https://doi.org/10.1038/nbt1097-984
Sambrook, J., & Russell, D. W. (2001). Molecular cloning: A laboratory manual. (3rd ed.). New York (US): Cold Spring Harbor Laboratory Pr. https://www.cabdirect.org/cabdirect/abstract/19901616061. Accessed 1 March 2021
SAS. (2013). SAS version 9.2 of the Statistical Analysis System for Windows. SAS Institute Inc.
Seo, J., Jeon, J., Kim, S.-D., Kang, S., Han, J., & Hur, H.-G. (2007). Fungal biodegradation of carbofuran and carbofuran phenol by the fungus Mucor ramannianus: Identification of metabolites. Water Sciences and Technology, 55(1–2), 163–167. https://doi.org/10.2166/wst.2007.051
Silambarasan, S., & Abraham, J. (2013). Ecofriendly method for bioremediation of chlorpyrifos from agricultural soil by novel fungus Aspergillus terreus JAS1. Water, Air, and Soil Pollution 224, 1369. https://doi.org/10.1007/s11270-012-1369-0
Singh, D. P., Khattar, J. I. S., Nadda, J., Singh, Y., Garg, A., Kaur, N., & Gulati, A. (2011). Chlorpyrifos degradation by the cyanobacterium Synechocystis sp. strain PUPCCC 64. Environmental Science and Pollution Research, 8(8), 1351–1359. https://doi.org/10.1007/s11356-011-0472-x
Solomon, K. R., Williams, W. M., Mackay, D., Purdy, J., Giddings, J. M., & Giesy, J. P. (2014). Properties and Uses of Chlorpyrifos in the United States. In: Giesy, J., Solomon, K. (eds) Ecological Risk Assessment for Chlorpyrifos in Terrestrial and Aquatic Systems in the United States. Reviews of Environmental Contamination and Toxicology, vol 231. Springer, Cham. https://doi.org/10.1007/978-3-319-03865-0_2
Subhas, & Singh, D. K. (2003). Utilization of monocrotophos as phosphorus source by Pseudomonas aeruginosa F10B and Clavibacter michiganense subsp. insidiosum SBL 11. Canadian Journal of Microbiology 49(2). https://doi.org/10.1139/w03-013
Supreeth, M., & Raju, N. (2017). Biotransformation of chlorpyrifos and endosulfan by bacteria and fungi. Applied Microbiology and Biotechnology 101: 5961–5971. https://doi.org/10.1007/s00253-017-8401-7
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30(12), 2725–2729. https://doi.org/10.1093/molbev/mst197
Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22(22), 4673–4680. https://doi.org/10.1093/NAR/22.22.4673
UN-WWDR. (2020). WWAP (UNESCO World Water Assessment Programme), 2019, United Nations World Water Development Report 2020: Water and Climate Change. https://reliefweb.int/report/world/worldwater-development-report-2020-water-and-climate-change-gclid=CjwKCAjw04yjBhApEiwAJcvNoRWmveYiTRwsxJNp5OwkVuxDGXzR8dJW3-2gHhjxoVqixMJr8THgbhoCM3EQAvD_BwE. Accessed Mar 2021.
Urovic, R., & Orevi, T. (2011). Modern extraction techniques for pesticide residues determination in plant and soil samples. Pesticides in the Modern World - Trends in Pesticides Analysis. InTech. https://doi.org/10.5772/17312
Vay, D., Parodi, M., Rolla, R., Mottaran, E., Vidali, M., Bellomo, G., & Albano, E. (2001). Circulating antibodies recognizing malondialdehyde-modified proteins in healthy subjects. Free Radical Biology and Medicine. 30(3): 277–286. https://doi.org/10.1016/S0891-5849(00)00469-X
Veeningen, C. (1999). National water resources plan volume II report. Hydro Delft. https://faolex.fao.org/docs/pdf/egy147082.pdf. Accessed Jun 2020.
Vidali, M. (2001). Bioremediation. An overview. Pure Applyed Chemistry 73(7): 1163–1172. https://doi.org/10.1351/pac200173071163
Vroumsia, T., Steinman, R., Seiglemurandi, F., & Benoitguyod, J. (2005). Fungal bioconversion of 2,4-dichlorophenoxyacetic acid (2,4-D) and 2,4-dichlorophenol (2,4-DCP). Chemosphere, 60(10), 1471–1480. https://doi.org/10.1016/j.chemosphere.2004.11.102
Xu, G., Li, Y., Zheng, W., Peng, X., Li, W., & Yan, Y. (2007). Mineralization of chlorpyrifos by co-culture of Serratia and Trichosporon spp. Biotechnology Letters 29: 1469–1473. https://doi.org/10.1007/s10529-007-9444-0
Yang, L., Zhao, Y. H., Zhang, B. X., Yang, C. H., & Zhang, X. (2005). Isolation and characterization of a chlorpyrifos and 3,5,6-trichloro-2- pyridinol degrading bacterium. FEMS Microbiology Letters 251(1):67–73. https://doi.org/10.1016/j.femsle.2005.07.031
Yu, Y. L., Fang, H., Wang, X., Wu, X. M., Shan, M., & Yu, J. Q. (2006). Characterization of a fungal strain capable of degrading chlorpyrifos and its use in detoxification of the insecticide on vegetables. Biodegradation 17: 487–494 . https://doi.org/10.1007/s10532-005-9020-z
Acknowledgements
Fungal species identification was completed with the help of Prof. Mohamed Magdi, Microbiology Department, Faculty of Science, Damanhour University.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
Atef Nassar and Sabha El-Sabbagh contributed to the study’s conception and design. Atef Nassar, Eman Nour Eldeen, and Eman Elzakey completed the laboratory experiments, Ibrahim Adss conducted the molecular identification of the bacterial strains, and Atef Nassar wrote and submitted the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
No ethical issue is to be declared in this article.
Consent to participate
No consent of participation is to be claimed.
Consent for publication
All authors have read and approved the manuscript before being sent for publication. Also, all authors confirm that it has not been published previously nor being considered by any other peer-reviewed journal.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Elzakey, E.M., El-Sabbagh, S.M., Eldeen, E.ES.N. et al. Bioremediation of chlorpyrifos residues using some indigenous species of bacteria and fungi in wastewater. Environ Monit Assess 195, 779 (2023). https://doi.org/10.1007/s10661-023-11341-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-023-11341-3