Abstract
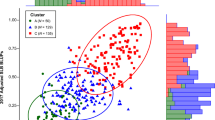
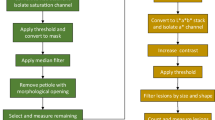
Fire blight, caused by Erwinia amylovora, is a destructive bacterial disease that severely hampers apple production. To conduct Quantitative Trait Locus (QTL) studies for breeding resistant apple cultivars, phenotyping of large genetic mapping populations of apples for fire blight resistance is essential. This, however, necessitates precise, quantitative data spanning multiple years, locations, and pathogen strains. It can be time-consuming and resource-intensive to keep QTL mapping populations for apples in the field and greenhouse. This creates a bottleneck for identifying novel QTL for fire blight resistance or developing resistant cultivars. To address this challenge, we present an image-based method for rapid and accurate phenotyping fire blight resistance using apple leaf discs. This leaf disc assay demonstrates significant (p < 0.05) percent disease area (PDA) differences in fire blight inoculations among eight apple genotypes with well-known resistance levels. Furthermore, the image-based leaf disc assay consistently shows a 40–70% difference in PDA between resistant and susceptible checks. We also report high within and across trial broad sense heritability values ranging from 0.86 – 0.97. We demonstrate the use of K-means clustering and best linear unbiased estimators to combine multiple trials. This assay offers an efficient alternative to traditional fire blight screening methods, potentially improving our understanding of the host response, and accelerating the development of resistant apple cultivars.




Similar content being viewed by others
References
Abdelraheem, A., Elassbli, H., Zhu, Y., Kuraparthy, V., Hinze, L., Stelly, D., Wedegaertner, T., & Zhang, J. (2020). A genome-wide association study uncovers consistent quantitative trait loci for resistance to Verticillium wilt and Fusarium wilt race 4 in the US Upland cotton. Theoretical and Applied Genetics, 133(2), 563–577. https://doi.org/10.1007/S00122-019-03487-X/FIGURES/3
Bates, D., Maechler, M., Bolker, B., Walker, S., Christensen, R. H. B., Singmann, H., Dai, B., Scheipl, F., Grothendieck, G. & Green, P. (2015). Package ‘lme4’: linear mixed-effects models using Eigen and S4 (Version 1.1-7). R Foundation for Statistical Computing. Available online at https://cran.r-project.org/web/packages/lme4/lme4.pdf. Assessed on 07 April 2023.
Bedford, K. E., Sholberg, P. L., & Kappel, F. (2003). Use of a detached leaf bioassay for screening sweet cherry cultivars for bacterial canker resistance. Acta Horticulturae, 622, 365–368. https://doi.org/10.17660/ACTAHORTIC.2003.622.37
Bernardo, R. (2020). Reinventing quantitative genetics for plant breeding: something old, something new, something borrowed, something BLUE. Heredity, 1256. 125:375–385. https://www.nature.com/articles/s41437-020-0312-1
Bock, C. H., Parker, P. E., Cook, A. Z., and Gottwald, T. R. 2008. Visual rating and the use of image analysis for assessing different symptoms of citrus canker on grapefruit leaves. Plant Disease, 92:530–541. https://pubmed.ncbi.nlm.nih.gov/30769647/
Bock, C. H., Poole, G. H., Parker, P. E., and Gottwald, T. R. (2010). Plant Disease Severity Estimated Visually, by Digital Photography and Image Analysis, and by Hyperspectral Imaging. Critical Reviews in Plant Sciences, 29(2), 59–107. https://www.tandfonline.com/doi/abs/https://doi.org/10.1080/07352681003617285
Bock, C. H., Chiang, K. S., and Del Ponte, E. M. (2022). Plant disease severity estimated visually: a century of research, best practices, and opportunities for improving methods and practices to maximize accuracy. Tropical Plant Pathology, 47:25–42. https://link.springer.com/article/https://doi.org/10.1007/s40858-021-00439-z
Brachi, B., Morris, G. P., Borevitz, J. O. (2011). Genome-wide association studies in plants: the missing heritability is in the field. Genome Biology, 12, 232. https://doi.org/10.1186/gb-2011-12-10-232
Calenge, F., Drouet, D., Denancé, C., Van De Weg, W. E., Brisset, M. N., Paulin, J. P., et al. (2005). Identification of a major QTL together with several minor additive or epistatic QTLs for resistance to fire blight in apple in two related progenies. Theoretical Applied Genetics, 111:128–135. https://link.springer.com/article/https://doi.org/10.1007/s00122-005-2002-z
Chiang, K. S., Bock, C. H., Lee, I. H., El Jarroudi, M., and Delfosse, P. (2016). Plant disease severity assessment-how rater Bias, assessment method, and experimental design affect hypothesis testing and resource use efficiency. Phytopathology, 106,1451–1464. https://apsjournals.apsnet.org/doi/https://doi.org/10.1094/PHYTO-12-15-0315-R
Cowger, C., and Brown, J. K. M. (2019). Durability of Quantitative Resistance in Crops: Greater Than We Know? Annual Review Phytopathology, 57, 253–277. https://pubmed.ncbi.nlm.nih.gov/31206351/
Desnoues, E., Norelli, J. L., Aldwinckle, H. S., Wisniewski, M. E., Evans, K. M., Malnoy, M., et al. (2018). Identification of Novel Strain-Specific and Environment-Dependent Minor QTLs Linked to Fire Blight Resistance in Apples. Plant Molecular Biology Report, 36, 247–256. https://link.springer.com/article/https://doi.org/10.1007/s11105-018-1076-0
Donovan, A. (1991). Screening for fire blight resistance in apple (Malus pumila) using excised leaf assays from in vitro and in vivo grown material. Annual Applied Biology, 119, 59–68 https://onlinelibrary.wiley.com/doi/full/https://doi.org/10.1111/j.1744-7348.1991.tb04843.x
Durel, C. E., Denancé, C., and Brisset, M. N. (2009). Two distinct major QTL for resistance to fire blight co-localize on linkage group 12 in apple genotypes “Evereste” and Malus floribunda clone 821. Genome, 52,139–147 https://cdnsciencepub.com/doi/https://doi.org/10.1139/G08-111
Emeriewen, O. F., Richter, K., Flachowsky, H., Malnoy, M., and Peil, A. (2021). Genetic Analysis and Fine Mapping of the Fire Blight Resistance Locus of Malus × arnoldiana on Linkage Group 12 Reveal First Candidate Genes. Frontiers in Plant Science, 12, 663. https://www.frontiersin.org/articles/https://doi.org/10.3389/fpls.2021.667133/full
Fox, J. (2007). The car package. R Foundation for Statistical Computing. https://rdocumentation.org/packages/car/versions/3.1-1
Fox, J., Weisberg, S., Price, B., Adler, D., Bates, D., Baud-Bovy, G. & Bolker, B. (2019). car: Companion to Applied Regression. R package version 3.0-2. Website. https://CRAN.R-project.org/package=car. Accessed 24 April 2023.
Harshman, J. M., Evans, K. M., Allen, H., Potts, R., Flamenco, J., Aldwinckle, H. S., et al. (2017). Fire blight resistance in wild accessions of Malus sieversii. Plant Disease, 101, 1738–1745. https://apsjournals.apsnet.org/doi/https://doi.org/10.1094/PDIS-01-17-0077-RE
Kassambara A, Mundt F. (2017). Package ‘factoextra’. Extract and visualize the results of multivariate data analyses. http://www.sthda.com/english/rpkgs/factoextra.
Khan, A., and Korban, S. S. (2022). Breeding and genetics of disease resistance in temperate fruit trees: challenges and new opportunities. Theoretical Applied Genetics, 13511, 135:3961–3985. https://link.springer.com/article/https://doi.org/10.1007/s00122-022-04093-0
Khan, M. A., Duffy, B., Gessler, C., and Patocchi, A. (2006). QTL mapping of fire blight resistance in apple. Molecular Breeding, 17, 299–306. https://link.springer.com/article/https://doi.org/10.1007/s11032-006-9000-y
Khan, M. A., Durel, C. E., Duffy, B., Drouet, D., Kellerhals, M., Gessler, C., et al. (2007). Development of molecular markers linked to the “Fiesta” linkage group 7 major QTL for fire blight resistance and their application for marker-assisted selection. Genome, 50, 568–577. https://cdnsciencepub.com/doi/https://doi.org/10.1139/G07-033
Kharadi, R. R., Schachterle, J. K., Yuan, X., Castiblanco, L. F., Peng, J., Slack, S. M., Zeng, Q., & Sundin, G. W. (2021). Genetic Dissection of the Erwinia amylovora Disease Cycle. Annual Review Phytopathology, 191–212. https://doi.org/10.1146/ANNUREV-PHYTO-020620-095540
Kortekamp, A. (2006). Expression analysis of defense-related genes in grapevine leaves after inoculation with a host and a non-host pathogen. Plant Physiology and Biochemistry, 44(1), 58–67. https://doi.org/10.1016/J.PLAPHY.2006.01.008
Kostick, S. A., Norelli, J. L., and Evans, K. M. (2019). Novel metrics to classify fire blight resistance of 94 apple cultivars. Plant Pathology, 68, 985–996. https://onlinelibrary.wiley.com/doi/full/https://doi.org/10.1111/ppa.13012
Kostick, S. A., Norelli, J. L., Teh, S. L., and Evans, K. M. (2021). Quantitative variation and heritability estimates of fire blight resistance in a pedigree-connected apple germplasm set. J. Plant Pathology, 103, 65–75. https://link.springer.com/article/https://doi.org/10.1007/s42161-020-00543-0
Kruijer, W., Boer, M. P., Malosetti, M., Flood, P. J., Engel, B., Kooke, R., Keurentjes, J. J. B., & van Eeuwijk, F. A. (2014). Marker-based estimation of heritability in immortal populations. Genetics, 199(2), 379–398. https://doi.org/10.1534/GENETICS.114.167916/-/DC1
Leonards-Schippers, C., Gieffers, W., Schafer-Pregl, R., Ritter, E., Knapp, S. J., Salamini, F., et al. (1994). Quantitative resistance to Phytophthora infestans in potato: a case study for QTL mapping in an allogamous plant species. Genetics, 137, 67–77. https://academic.oup.com/genetics/article/137/1/67/6012520
Li, L., Zhang, Q., and Huang, D. (2014). A review of imaging techniques for plant phenotyping. Sensors (Switzerland), 14, 20078–20111. https://www.mdpi.com/1424-8220/14/11/20078
Longzhou, L., Xiaojun, Y., Run, C., Junsong, P., Huanle, H., Lihua, Y., Yuan, G., & Lihuang, Z. (2008). Quantitative trait loci for resistance to powdery mildew in cucumber under seedling spray inoculation and leaf disc infection. Journal of Phytopathology, 156(11–12), 691–697. https://doi.org/10.1111/j.1439-0434.2008.01427.x
Martínez-Bilbao, A., Ortiz-Barredo, A., Montesinos, E., and Murillo, J. S. (2009). Evaluation of a Cider Apple Germplasm Collection of Local Cultivars from Spain for Resistance to Fire Blight (Erwinia amylovora) Using a Combination of Inoculation Assays on Leaves and Shoots. Horticulture Science, 44, 1223–1227 https://journals.ashs.org/hortsci/view/journals/hortsci/44/5/article-p1223.xml
Mir, R. R., Reynolds, M., Pinto, F., Khan, M. A., and Bhat, M. A. (2019). High-throughput phenotyping for crop improvement in the genomics era. Plant Science, 282, 60–72. https://pubmed.ncbi.nlm.nih.gov/31003612/
Moragrega, C., Llorente, I., Manceau, C., & Montesinos, E. (2003). Susceptibility of European pear cultivars to Pseudomonas syringae pv. syringae using immature fruit and detached leaf assays. European Journal of Plant Pathology, 109(4), 319–326. https://doi.org/10.1023/A:1023574219069/METRICS
Mutka, A. M., and Bart, R. S. (2015). Image-based phenotyping of plant disease symptoms. Front. Plant Science, 5, 734. https://www.frontiersin.org/articles/https://doi.org/10.3389/fpls.2014.00734/full
Panter, S. N., & Jones, D. A. (2002). Age-related resistance to plant pathogens. Advances in Botanical Research, 38. https://doi.org/10.1016/s0065-2296(02)38032-7
Peil, A., Garcia-Libreros, T., Richter, K., Trognitz, F. C., Trognitz, B., Hanke, M. V., & Flachowsky, H. (2007). Strong evidence for a fire blight resistance gene of Malus robusta located on linkage group 3. Plant Breeding, 126(5), 470–475. https://doi.org/10.1111/j.1439-0523.2007.01408.x
Peil, A., Hanke, M. V., Flachowsky, H., Richter, K., Garcia-Libreros, T., Celton, J. M., et al. (2008). Confirmation of the fire blight QTL of Malus × robusta 5 on linkage group 3. Acta Horticulturae, 793, 297–304. https://www.actahort.org/books/793/793_44.htm
Peil, A., Hübert, C., Wensing, A., Horner, M., Emeriewen, O. F., Richter, K., et al. (2019). Mapping of fire blight resistance in Malus × robusta 5 flowers following artificial inoculation. BMC Plant Biology, 19, 1–12. https://bmcplantbiol.biomedcentral.com/articles/https://doi.org/10.1186/s12870-019-2154-7
Peil, A., Emeriewen, O. F., Khan, A., Kostick, S., and Malnoy, M. (2021). Status of fire blight resistance breeding in Malus. Journal of Plant Pathology, 103, 3–12. https://link.springer.com/article/https://doi.org/10.1007/s42161-020-00581-8
Perez, L. B., & Brown, P. J. (2014). The role of ROS signaling in cross-tolerance: From model to crop. Frontiers in Plant Science, (DEC), 1–6. https://doi.org/10.3389/FPLS.2014.00754/BIBTEX
R Core Team. (2021). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.r-project.org/
R Core Team. (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.R-project.org/
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature Methods, 9, 676–682 https://www.nature.com/articles/nmeth.2019
Wallin, J., Bogdan, M., Szulc, P. A., Doerge, R. W., and Siegmund, D. O. (2021). Ghost QTL and hotspots in experimental crosses: novel approach for modeling polygenic effects. Genetics. https://academic.oup.com/genetics/article/217/3/iyaa041/6067404
Wang, P., Yin, L., Liang, D., Li, C., Ma, F., & Yue, Z. (2012). Delayed senescence of apple leaves by exogenous melatonin treatment: Toward regulating the ascorbate–glutathione cycle. Journal of Pineal Research, 53(1), 11–20. https://doi.org/10.1111/J.1600-079X.2011.00966.X
van de Weg, E., Di Guardo, M., Jänsch, M., Socquet-Juglard, D., Costa, F., Baumgartner, I., et al. (2018). Epistatic fire blight resistance QTL alleles in the apple cultivar ‘Enterprise’ and selection X-6398 discovered and characterized through pedigee-informed analysis. Molecular Breeding, 38, 1–18. https://link.springer.com/article/https://doi.org/10.1007/s11032-017-0755-0
Zendler, D., Malagol, N., Schwandner, A., Töpfer, R., Hausmann, L., and Zyprian, E. (2021). High-Throughput Phenotyping of Leaf Discs Infected with Grapevine Downy Mildew Using Shallow Convolutional Neural Networks. Agronomy, 11, 1768 https://www.mdpi.com/2073-4395/11/9/1768/htm
Acknowledgements
This research was funded by the New York State Department of Agriculture & Markets, Apple Research & Development Program (ARDP), grant number: CM04068AQ, Khan. We would like to acknowledge the USDA (United States Department of Agriculture) Plant Genetic Resources Unit (PGRU) in Geneva, New York, for maintaining the US national Malus collection, where all plant material was collected.
Author information
Authors and Affiliations
Contributions
A.K., conceptualized, designed, and managed the project. R.T., and D.H., done the experiments, analysis and interpretation. R.T., drafted the manuscript. R.T., and D.H., K.R., and A.K., revised, and finalized the manuscript. All authors read and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tegtmeier, R., Hickok, D., Robbins, K. et al. An image-analysis based leaf disc assay for the rapid evaluation of genetic resistance to fire blight in apples. Eur J Plant Pathol 168, 249–259 (2024). https://doi.org/10.1007/s10658-023-02750-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-023-02750-8