Abstract
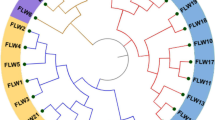
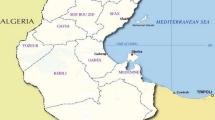
Powdery mildew, caused by Oidium heveae, is a major threat to rubber plantations worldwide. Population studies are scarce for this pathogen due to the lack of polymorphic molecular markers. In this study, sixteen polymorphic microsatellite loci were identified using DNA extracted from single lesions based on the whole-genome sequences of the pathogen. Primers of these loci were applied to 138 O. heveae samples from five counties in Hainan, China. The number of alleles per locus ranged from 2 to 11 and the gene diversity varied from 0.014 to 0.728. In total, 119 multilocus genotypes (MLGs) were observed for the 138 samples. However, only one sample was significantly distinct from the other samples base on DAPC analysis. Further study indicated that there were three subclusters for the other 137 samples, but no evidence for regional genotypic subdivision was identified. The microsatellite markers developed are very useful to study the genetic structure and the dispersal route of O. heveae, especially as it can use DNA extracted directly from infected leaves.




Similar content being viewed by others
References
Ali, S., Gautier, A., Leconte, M., Enjalbert, J., & de Vallavieille-Pope, C. (2011). A rapid genotyping method for an obligate fungal pathogen, Puccinia striiformis f. sp. tritici, based on DNA extraction from infected leaf and multiplex PCR genotyping. BMC Research Notes, 4, 240.
Ali, S., Gladieux, P., Leconte, M., Gautier, A., Justesen, A. F., Hovmøller, M. S., Enjalbert J., & Vallavieille, C. (2014) Origin, migration routes and worldwide population genetic structure of the wheat yellow rust pathogen Puccinia striiformis f.sp. tritici. PLoS Pathogens, 10(1), e1003903.
Benson, G. (1999). Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Research, 27(2), 573–580.
Chen, X. M., Chen, H. L., Li, W. G., & Liu, S. J. (2016). Remote sensing monitoring of spring phenophase of natural rubber forest in Hainan province. Chinese Journal of Agrometeorology, 37(1), 111–116.
Frenkel, O., Portillo, I., Brewer, M. T., Pèros, J. P., Cadle-Davidson, L., & Milgroom, M. G. (2012). Development of microsatellite markers from the transcriptome of Erysiphe necator for analyzing population structure in North America and Europe. Plant Pathology, 61, 106–119.
Glawe, D. A. (2008). The powdery mildews: A review of the world’s most familiar (yet poorly known) plant pathogens. Annual Review of Phytopathology, 46, 27–51.
Grover, A., & Sharma, P. C. (2016). Development and use of molecular markers: Past and present. Critical Reviews in Biotechnology, 36(2), 290–302.
Grünwald, N. J., Everhart, S. E., Knaus, B. J., & Kamvar, Z. N. (2017). Best practices for population genetic analyses. Phytopathology, 107(9), 1000–1010.
Jombart, T., Devillard, S., & Balloux, F. (2010). Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genetics, 11, 94.
Kamvar, Z. N., Tabima, J. F., & Grünwald, N. J. (2014). Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. Peer J, 2, e281.
Khan, M. R., Rehman, Z. U., Nazir, S. N., Tshewang, S., & Ali, S. (2019). Genetic divergence and diversity in himalayan Puccinia striiformis populations from Bhutan, Nepal, and Pakistan. Phytopathology, 109(10), 1793–1800.
Lederson, G. B., Peever, T. L., Evans, K., & Amiri, A. (2021). High genetic diversity in predominantly clonal populations of the powdery mildew fungus Podosphaera leucotricha from U.S. apple orchards. Applied and Environmental Microbiology, 87(15), e0046921.
Limkaisang, S., Komun, S., Furtado, E. L., Liew, K. W., Salleh, B., Sato, Y., & Takamatsu, S. (2005). Molecular phylogenetic and morphological analyses of Oidium heveae, a powdery mildew of rubber tree. Mycoscience, 46, 220–226.
Liu, S. J., Zhou, G. S., & Fang, S. B. (2015). Climatic suitability regionalization of rubber plantation in China. Scientia Agricultura Sinica, 48, 2335–2345.
Liyanage, K. K., Khan, S., Mortimer, P. E., Hyde, K. D., Xu, J., Brooks, S., & Ming, Z. (2016). Powdery mildew disease of rubber tree. Forest Pathology, 46, 90–103.
Liyanage, K. K., Khan, S., Brooks, S., Mortimer, P. E., Karunarathna, S. C., Xu, J., & Hyde, K. D. (2017). Taxonomic revision and phylogenetic analyses of rubber powdery mildew fungi. Microbial Pathogenesis, 105, 185–195.
Maddalena, G., Delmotte, F., Bianco, P. A., Lorenzis, G. D., & Toffolatti, S. L. (2020). Genetic structure of italian population of the grapevine downy mildew agent, plasmopara viticola. Annals of Applied Biology, 176(3), 257–267.
Nicolaisen, M., West, J. S., Sapkota, R., Canning, G. G. M., Schoen, C., & Justesen, A. F. (2017). Fungal communities in near surface air are similar across northwestern Europe. Frontiers in Microbiology, 8, 1729.
Qin, Y. X., Zhou, H. Z., Chen, J., Yang, J. H., Hu, Y. S., & Tang, C. R. (2017). Predominant ontamination agents in rubber tree tissue culture and its elimination. Botanical Research, 6(3), 167–174.
Szpiech, Z. A., Jakobsson, M., & Rosenberg, A. N. A. (2008). Adze: A rarefaction approach for counting alleles private to combinations of populations. Bioinformatics, 24(21), 2498–2504.
Tam, L. T. T., Dung, P. N., & Liem, N. V. (2016). First report of powdery mildew caused by Erysiphe quercicola on mandarin in Vietnam. Plant Disease, 100, 1777.
Tu, M., Cai, H. B., Hua, Y. W., Sun, A. H., & Huang, H. S. (2012). In vitro culture method of powdery mildew (Oidium heveae Steinmann) of Hevea brasiliensis. African Journal of Biotechnology, 11(68), 13167–13172.
Tucker, M. A., Moffat, C. S., Ellwood, S. R., Tan, K. C., & Oliver, R. P. (2015). Development of genetic SSR markers in Blumeria graminis f. sp. hordei and application to isolates from Australia. Plant Pathology, 64(2), 337–343.
Wang, M., Xue, F., Yang, P., Duan, X. Y., Zhou, Y. L., Sheng, C. Y., Zhang, G. Z., & Wang, B. T. (2014). Development of SSR markers for a phytopathogenic fungus, Blumeria graminis f.sp. tritici, using a fiasco protocol. Journal of Integrative Agriculture, 13(1), 100–104.
Wu, H., Pan, Y., Di, R., He, Q., Rajaofera, M. J. N., Liu, W., & Miao, W. (2019). Molecular identification of the powdery mildew fungus infecting rubber trees in China. Forest Pathology, 49, e12519.
Wyman, C. R., Hadziabdic, D., Boggess, S. L., Rinehart, T. A., & Trigiano, R. N. (2019). Low genetic diversity suggests the recent introduction of dogwood powdery mildew to North America. Plant Disease, 103(11), 2903–2912.
Funding
This work was funded by the National Natural Science Foundation of China (31972212, 31701731) and Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (No. 1630042017003). The authors have no conflict of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Supplementary Information
Fig. S1
The genotype accumulation curve for 138 rubber tree powdery mildew samples from Hainan, China. The boxplots represent the range of MLG numbers observed for each random sample of loci. (PNG 616 kb)
Fig. S2
Discriminant analysis of principal components (DAPC) of the 137 rubber tree powdery mildew samples from Hainan, China. A, Value of Bayesian information criteria (BIC); B, Scatterplot of DAPC. (PNG 75.9 kb)
ESM 1
(XLS 23.3 kb)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Han, Q., He, Y., Che, H. et al. Development and characterization of microsatellite markers for the rubber tree powdery mildew pathogen Oidium heveae. Eur J Plant Pathol 164, 253–262 (2022). https://doi.org/10.1007/s10658-022-02555-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-022-02555-1