Abstract
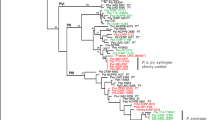
Bacterial wilt caused by Curtobacterium flaccumfaciens pv. flaccumfaciens is among the diseases that affect Phaseolus vulgaris L. This disease has been frequently detected in bean fields and causes severe production losses in Brazil. The aim of this research was to examine the genetic diversity existing among twenty-four isolates of C. flaccumfaciens collected from their native and alternative host, and a collection of sixty strains belonging to four phytopathogenic pathovars preserved at the French Collection for Plant-associated Bacteria (CIRM-CFBP) by multilocus sequence analysis (MLSA) based on six housekeeping genes (atpD, dnaK, gyrB, ppK, recA and rpoB). A phylogenetic tree with the concatenated sequences of six genes showed high genetic diversity among the strains. For instance, strains belonging to C. f. pv. flaccumfaciens do not cluster together within the species. Similar results were obtained with a minimal MLSA scheme using gyrB and recA, which we propose for reliable identification at the species level of Curtobacterium isolates. No correlation was identified between phylogeny and pathogenicity in the Curtobacterium flaccumfaciens strains analyzed in this work. The specific primers CffFOR2 and CffREV4 designed by Tegli et al. (Letters in Applied Microbiology, 35(4), 331–337, 2002) to detect C. f. pv. flaccumfaciens in naturally infected bean seeds proved to be efficient for the detection of bean-pathogenic strains.


Similar content being viewed by others
References
Agarkova, I. V., Lambrecht, P. A., Vidaver, A. K., & Harveson, R. M. (2012). Genetic diversity among Curtobacterium flaccumfaciens pv. flaccumfaciens populations in the American High Plains. Canadian Journal of Microbiology, 58(6), 788–801. https://doi.org/10.1139/w2012-052.
Bishop, C. J., Aanensen, D. M., Jordan, G. E., Kilian, M., Hanage, W. P., & Spratt, B. G. (2009). Assigning strains to bacterial species via the internet. BMC Biology, 7(1), 3. https://doi.org/10.1186/1741-7007-7-3.
Chen, Y. F., Guo, J. H., & Fang, Z. D. (2000). A new pathovar of Curtobacterium flaccumfaciens on Malabar spinach. Acta Phytopathologica Sinica, 30, 171–175.
Chen, Y.-F., Yin, Y.-N., Zhang, X.-M., & Guo, J.-H. (2007). Curtobacterium flaccumfaciens pv. beticola , a new pathovar of pathogens in sugar beet. Plant Disease, 91(6), 677–684. https://doi.org/10.1094/PDIS-91-6-0677.
Collins, M. D., & Jones, D. (1983). Reclassification of Corynebacterium flaccumfaciens, Corynebacterium betae, Corynebacterium oortii and Corynebacterium poinsettiae in the genus Curtobacterium, as Curtobacterium flaccumfaciens comb. nov. Microbiology, 129(11), 3545–3548. https://doi.org/10.1099/00221287-129-11-3545.
Darsonval, A., Darrasse, A., Durand, K., Bureau, C., Cesbron, S., & Jacques, M.-A. (2009). Adhesion and fitness in the bean phyllosphere and transmission to seed of Xanthomonas fuscans subsp. fuscans. Molecular Plant-Microbe Interactions, 22(6), 747–757. https://doi.org/10.1094/MPMI-22-6-0747.
de Souza, V. L., Maringoni, A. C., & Krause-Sakate, R. (2006). Genetic variability in Curtobacterium flaccumfaciens isolates. Summa Phytopathologica, 32(2), 170–176. https://doi.org/10.1590/S0100-54052006000200012.
Dow, J. M., Clarke, B. R., Milligan, D. E., Tang, J. L., & Daniels, M. J. (1990). Extracellular proteases from Xanthomonas campestris pv. campestris, the black rot pathogen. Applied and Environmental Microbiology, 56(10), 2994–2998.
Fischer-Le Saux, M., Bonneau, S., Essakhi, S., Manceau, C., & Jacques, M.-A. (2015). Aggressive emerging pathovars of Xanthomonas arboricola represent widespread epidemic clones distinct from poorly pathogenic strains, as revealed by multilocus sequence typing. Applied and Environmental Microbiology, 81(14), 4651–4668. https://doi.org/10.1128/AEM.00050-15.
Gonçalves, R. M., Schipanski, C. A., Koguishi, L., Soman, J. M., Sakate, R. K., Júnior, T. A. F. S., & Maringoni, A. C. (2017). Alternative hosts of Curtobacterium flaccumfaciens pv. flaccumfaciens, causal agent of bean bacterial wilt. European Journal of Plant Pathology, 148(2), 357–365. https://doi.org/10.1007/s10658-016-1094-4.
Guimarães, P. M., Smith, J. J., Palmano, S., & Saddler, G. S. (2003). Characterisation of Curtobacterium flaccumfaciens pathovars by AFLP, rep-PCR and pulsed-field gel electrophoresis. European Journal of Plant Pathology, 109(8), 817–825. https://doi.org/10.1023/A:1026197914417.
Hajri, A., Pothier, J. F., Fischer-Le Saux, M., Bonneau, S., Poussier, S., Boureau, T., et al. (2012). Type three effector gene distribution and sequence analysis provide new insights into the pathogenicity of plant-pathogenic Xanthomonas arboricola. Applied and Environmental Microbiology, 78(2), 371–384. https://doi.org/10.1128/AEM.06119-11.
Hedges, F. (1922). A bacterial wilt of the bean caused by Bacterium flaccumfaciens nov. sp. Science, 55(1425), 433–434. https://doi.org/10.1126/science.55.1425.433.
Hedges, F. (1926). Bacterial wilt of beans (Bacterium flaccumfaciens Hedges), including comparisons with Bacterium phaseoli. Phytopathology, 16(1), 1–22.
Herbes, D. H., Theodoro, G. F., Maringoni, A. C., dal Piva, C. A., & de Abreu, L. (2008). Detection of Curtobacterium flaccumfaciens pv. flaccumfaciens in seeds of common bean produced in Santa Catarina. Tropical Plant Pathology, 33(2), 153–156. https://doi.org/10.1590/S1982-56762008000200010.
Huson, D. H., & Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution, 23(2), 254–267.
Jacques, M. A., Durand, K., Orgeur, G., Balidas, S., Fricot, C., Bonneau, S., Quillévéré, A., Audusseau, C., Olivier, V., Grimault, V., & Mathis, R. (2012). Phylogenetic analysis and polyphasic characterization of Clavibacter michiganensis strains isolated from tomato seeds reveal that nonpathogenic strains are distinct from C. michiganensis subsp. michiganensis. Applied and Environmental Microbiology, 78(23), 8388–8402. https://doi.org/10.1128/AEM.02158-12.
Li, R., Zhu, H., Ruan, J., Qian, W., Fang, X., Shi, Z., Li, Y., Li, S., Shan, G., Kristiansen, K., Li, S., Yang, H., Wang, J., & Wang, J. (2010). De novo assembly of human genomes with massively parallel short read sequencing. Genome Research, 20(2), 265–272. https://doi.org/10.1101/gr.097261.109.
Maringoni, A. C. (2002). Behaviour of dry bean cultivars to bacterial wilt. Fitopatologia Brasileira, 27(2), 157–162. https://doi.org/10.1590/S0100-41582002000200006.
Maringoni, A. C., & Rosa, E. F. (1997). Occurrence of Curtobacterium flaccumfaciens pv. flaccumfaciens on bean in the state of Sao Paulo, Brazil. Summa Phytopathologica (Brazil), 23, 160–162.
Merda, D., Briand, M., Bosis, E., Rousseau, C., Portier, P., Barret, M., Jacques, M. A., & Fischer-le Saux, M. (2017). Ancestral acquisitions, gene flow and multiple evolutionary trajectories of the type three secretion system and effectors in Xanthomonas plant pathogens. Molecular Ecology, 26(21), 5939–5952. https://doi.org/10.1111/mec.14343.
Osdaghi, E., Taghavi, S. M., Hamzehzarghani, H., Fazliarab, A., Harveson, R. M., & Lamichhane, J. R. (2016). Occurrence and characterization of a new red-pigmented variant of Curtobacterium flaccumfaciens, the causal agent of bacterial wilt of edible dry beans in Iran. European Journal of Plant Pathology, 146(1), 129–145. https://doi.org/10.1007/s10658-016-0900-3.
Osdaghi, E., Taghavi, S. M., Hamzehzarghani, H., Fazliarab, A., Harveson, R. M., Tegli, S., & Lamichhane, J. R. (2018a). Epiphytic Curtobacterium flaccumfaciens strains isolated from symptomless solanaceous vegetables are pathogenic on leguminous but not on solanaceous plants. Plant Pathology, 67, 388–398. https://doi.org/10.1111/ppa.12730.
Osdaghi, E., Taghavi, S. M., Calamai, S., Biancalani, C., Cerboneschi, M., Tegli, S., & Harveson, R. (2018b). Phenotypic and molecular-phylogenetic analysis provide novel insights into the diversity of Curtobacterium flaccumfaciens. Phytopathology, 108, 1154–1164.
Sallet, E., Gouzy, J., & Schiex, T. (2014). EuGene-PP: A next-generation automated annotation pipeline for prokaryotic genomes. Bioinformatics, 30(18), 2659–2661. https://doi.org/10.1093/bioinformatics/btu366.
Sammer, U. F., & Reiher, K. (2012). Curtobacterium flaccumfaciens pv. flaccumfaciens on soybean in Germany - a threat for farming. Journal of Phytopathology, 160(6), 314–316. https://doi.org/10.1111/j.1439-0434.2012.01902.x.
Schwartz, H. F., Steadman, J. R., Hall, R., & Forster, R. L. (2005). Compendium of bean diseases. St. Paul: APS Press American Phytopathological Society.
Tancos, M. A., Lange, H. W., & Smart, C. D. (2015). Characterizing the genetic diversity of the Clavibacter michiganensis subsp. michiganensis population in New York. Phytopathology, 105(2), 169–179. https://doi.org/10.1094/PHYTO-06-14-0178-R.
Tegli, S., Sereni, A., & Surico, G. (2002). PCR-based assay for the detection of Curtobacterium flaccumfaciens pv. flaccumfaciens in bean seeds. Letters in Applied Microbiology, 35(4), 331–337.
Theodoro, G. d. F., Maringoni, A. C., Chumpati, A. A., Correia, H. d. C., Theodoro, J. V. C., & Nogueira, R. J. (2010). First report of bacterial wilt of common bean caused by Curtobacterium flaccumfaciens pv. flaccumfaciens in Mato Grosso do Sul. Journal of Plant Pathology, 92, S4.107–S4.122.
Uesugi, C. H., Freitas, M. A., & Menezes, J. R. (2003). First occurrence of Curtobacterium flaccumfaciens pv. flaccumfaciens on bean in the state of Goias and Federal District of Brazil. Fitopatologia Brasileira, 28(3), 324–324. https://doi.org/10.1590/S0100-41582003000300019.
Vignesh, R., Prabakar, P., Jaganathan, R., & Swathirajan, C. R. (2016). Isolation and characterization of extracellular proteases from Pseudomonas aeruginosa and Bacillus subtilis strains. Bull. Env. Pharmacol. Life Sci., 5(8), 40–43.
Young, J. M., Dye, D. W., Bradbury, J. F., Panagopoulos, C. G., & Robbs, C. F. (1978). A proposed nomenclature and classification for plant pathogenic bacteria. New Zealand Journal of Agricultural Research, 21(1), 153–177. https://doi.org/10.1080/00288233.1978.10427397.
Young, J. M., Watson, D. R. W., & Dye, D. W. (2004). Reconsideration of Arthrobacter ilicis (Mandel et al. 1961) Collins et al. 1982 as a plant-pathogenic species. Proposal to emend the authority and description of the species. Request for an opinion. International Journal of Systematic and Evolutionary Microbiology, 54(1), 303–305.
Zerbino, D. R., & Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Research, 18(5), 821–829. https://doi.org/10.1101/gr.074492.107.
Acknowledgments
The authors would like to thank CAPES (Coordination for the Improvement of Higher Education Personnel-Brazil) for financial support and Cécile Dutrieux, Karine Durand, and Armelle Darrasse for technical assistance and thoughtful discussions. The authors thank the reviewers of the first version of this paper for their very helpful comments, and Jason Shiller for the text review.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Supplemental Figure 1
(DOCX 144 kb)
Supplemental Figure 2
(DOCX 328 kb)
Supplemental Table 1
(XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Gonçalves, R.M., Balbi-Peña, M.I., Soman, J.M. et al. Genetic diversity of Curtobacterium flaccumfaciens revealed by multilocus sequence analysis. Eur J Plant Pathol 154, 189–202 (2019). https://doi.org/10.1007/s10658-018-01648-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-018-01648-0