Abstract
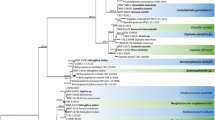
Leaves with typical symptoms of downy mildew were found on common agrimony in the Czech Republic in 2014 and 2015 and at several locations in Germany from 2010 to 2014. The causal agent of downy mildew of agrimony was often reported as Peronospora agrimoniae, but sometimes also as P. sparsa. Morphological characteristics of the pathogens found in both countries are in the range of previous works for P. agrimoniae, but also other downy mildews parasitic on Rosaceae, rendering their discrimination based on published observations difficult. For molecular identification sequencing of several loci (nrITS rDNA, cox1 and cox2) was performed. Phylogenetic analyses based on nrITS rDNA clearly separated P. agrimoniae from other Peronospora species infecting Rosaceae. Thus, considering P. agrimoniae as separate species seems justified. Two German specimens were identical to two Czech samples in both nrITS rDNA and cox1 mtDNA sequences, but differed in a single nucleotide substitution in cox2 region. To our knowledge, this is the first verified record of P. agrimoniae on common agrimony in the Czech Republic.



Similar content being viewed by others
References
Bonants, P., de Weerdt, M. H., van Gent Pelzer, M., Lacourt, I., Cooke, D., & Duncan, J. (1997). Detection and identification of Phytophtora fragariae Hickman by the polymerase chain reaction. European Journal of Plant Pathology, 103, 345–355.
Bontea, V. (1953). Ciuperci Parazite si Saprofite din Republica Populara Romana. Bucharest: Editura Academiei R. P. R.
Brandenburger, W., & Hagedorn, G. (2006). Zur Verbreitung von Peronosporales (inkl. Albugo, ohne Phytophthora) in Deutschland. Mitteilungen aus der Biologischen Bundesanstalt für Land– und Forstwirtschaft Berlin–Dahlem, 405, 1–174.
Choi, Y. J., Hong, S. B., & Shin, H. D. (2005). A reconsideration of Pseudoperonospora cubensis and Ps. humuli based on molecular and morphological data. Mycological Research, 109, 841–848.
Choi, Y. J., Constantinescu, O., & Shin, H. D. (2007). A new downy-mildew of the Rosaceae: Peronospora oblatispora sp. nov. (Chromista, Peronosporales). Nova Hedwigia, 85, 93–101.
Choi, Y. J., Shin, H. D., & Thines, M. (2009). Two novel Peronospora species are associated with recent reports of downy mildew on sages. Mycological Research, 113, 1340–1350.
Choi, Y. J., Beakes, G., Glockling, S., Kruse, J., Nam, B., Nigrelli, L., Ploch, S., Shin, H. D., Shivas, R. G., Telle, S., Voglmayr, H., & Thines, M. (2015a). Towards a universal barcode of oomycetes—a comparison of the cox1 and cox2 loci. Molecular Ecology Resources, 15, 1275–1288.
Choi, Y. J., Klosterman, S. J., Kummer, V., Voglmayr, H., Shin, H. D., & Thines, M. (2015b). Multi-locus tree and species tree approaches toward resolving a complex clade of downy mildews (Straminipila, Oomycota), including pathogens of beet and spinach. Molecular Phylogenetics and Evolution, 86, 24–34.
Constantinescu, O. (1991). An annotated list of Peronospora names. Thunbergia, 15, 1–110.
Constantinescu, O., & Negrean, G. (1983). Check-list of Romanian Peronosporales. Mycotaxon, 16, 537–556.
Cooke, D. E. L., Drenth, A., Duncan, J. M., Wagels, G., & Brasier, C. M. (2000). A molecular phylogeny of Phytophtora and related Oomycetes. Fungal Genetics and Biology, 30, 17–32.
De Vienne, D. M., Giraud, T., & Martin, O. C. (2007). A congruence index for testingtopological similarity between trees. Bioinformatics, 23, 3119–3124.
Dick, M. W. (2001). Straminipilous fungi: Systematics of the Peronosporomycetes, including accounts of the marine straminipilous protists, the plasmodiophorids and similar organisms. Dordrecht: Kluwer Academic Publishers.
Duke, J. A., Bogenschutz-Godwin, M. J., duCellier, J., & Duke, P. A. K. (2002). Handbook of medicinal herbs. Boca Raton: CRC Press.
Edgar, R. (2004). Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797.
Edwards, K., Johnstone, C., & Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Research, 19, 1349.
Farr, D. F., & Rossman, A. Y. (2015). Fungal databases, systematic mycology and microbiology laboratory, ARS, USDA. http://nt.ars-grin.gov/fungaldatabases/. Accessed 2 Jul 2015.
Francis, S. M. (1981). Peronospora sparsa. CMI Descriptions of Pathogenic Fungi and Bacteria, 690, 1–2.
Gäumann, E. A. (1923). Beiträge zu einer Monographie der Gattung Peronospora Corda. Zürich: Gebr. Fretz.
Gustavsson, A. (1959). Studies on Nordic Peronosporas. I. Taxonomic revision. Opera Botanica, 3, 1–271.
Hall, G. (1991). Peronospora potentillae. IMI Descriptions of Fungi and Bacteria, 1061, 1–2.
Hatfield, G. (2004). Encyclopedia of folk medicine: Old world and new world traditions. Santa Barbara: ABC-CLIO.
Hirilovich, I. S., & Lemeza, N. A. (2010). Species structure and distribution of micromycetes order Peronosporales in vicinities of geostation “West Beresina”. Proceedings of the National Academy of Science of Belarus, Biological Series 2010 N 4, 27–35.
Horst, R. K. (2001). Westcott’s plant disease handbook. Boston: Kluwer Academic Publishers.
Hruby, J. (1930). Beiträge zur Pilzflora Mährens und Schlesiens. Hedwigia, 69, 173–211.
Hudspeth, D. S. S., Nadler, S. A., & Hudspeth, M. E. S. (2000). A cox2 molecular phylogeny of the Peronosporomycetes. Mycologia, 92, 674–684.
Huelsenbeck, J. P., & Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics, 17, 754–755.
Iwata, Y. (1942). Specialization in Pseudoperonospora cubensis (Berk. et Curt.) Rostov. II. Comparative studies of the morphologies of the fungi from Cucumis sativus L. and Cucurbita moschata Duchesne. Annals of the Phytopathological Society of Japan, 11, 172–185.
Jermalaviciute, S. (1962). Nauja medziaga Lietuvos TSR Peronosporainiams grybams (Peronosporales) pazinti. Lietuvos TSR Mokslu Akademija, Darbai Ser. C, 3(29), 13–22.
Jung, T., & Burgess, T. I. (2009). Re-evaluation of Phytophthora citricola isolates from multiple woody hosts in Europe and North America reveals a new species. Phytophthora plurivora sp. nov. Persoonia, 22, 95–110.
Kitner, M., Lebeda, A., Sharma, R., Runge, F., Dvořák, P., Tahir, A., Choi, Y. J., Sedláková, B., & Thines, M. (2015). Coincidence of virulence shifts and population genetic changes of Pseudoperonospora cubensis in the Czech Republic. Plant Pathology, 64, 1461–1670.
Kruse, J. (2014). Diversität der pflanzenpathogenen Kleinpilze im Ökologisch-Botanischen Garten der Universität Bayreuth. Zeitschrift für Mykologie, 80, 169–226.
Kućmierz, J. (1966). Parasitic fungi of the Ojcow National Park. Part II. Archimycetes, Phycomycetes, Ustilaginales. Fragmenta Floristica et Geobotanica, 12, 497–511.
Martin, F. N., & Tooley, P. W. (2003). Phylogenetic relationships among Phytophthora species inferred from sequence analysis of mitochondrially encoded cytochrome oxidase I and II genes. Mycologia, 95, 269–284.
Mishra, B., & Thines, M. (2014). siMBa—a simple graphical user interface for the Bayesian inference program MrBayes. Mycological Progress, 13, 1255–1258.
Moncalvo, J. M., Wang, H. H., & Hseu, R. S. (1995). Phylogenetic relationships in Ganoderma inferred from the internal transcribed spacers and 25S ribosomal DNA sequences. Mycologia, 87, 223–238.
Mordue, J. E. M. (1989). Peronospora rubi. CMI Descriptions of Pathogenic Fungi and Bacteria, 976, 1–2.
Mulenko, W., Majewski, T., & Ruszkiewicz-Michalska, M. (2008). Preliminary checklist of micromycetes in Poland. Biodiversity of Poland 9. Krakow: W. Szafer Institute of Botany, Polish Academy of Sciences.
Müller, J., & Kokeš, P. (2008). Extended checklist of downy mildews of Moravia and Czech Silesia. Czech Mycology, 60, 91–104.
Petrželová, I., Dušek, K., & Dušková, E. (2013). Monitoring of pests and diseases in the field collections of medicinal, aromatic and culinary plants (MAPs)—preliminary results. Olomouc Biotech 2013 Plant Biotechnology: Green for Good II, Book of Abstracts, 93.
Pirondi, A., Kitner, M., Iotti, M., Sedláková, B., Lebeda, A., & Collina, M. (2016). Genetic structure and phylogeny of Italian and Czech populations of the cucurbit powdery mildew fungus Golovinomyces orontii inferred by Multilocus Sequence Typing. Plant Pathology, 65, 959–967.
Robideau, G. P., de Cock, A. W. A. M., Coffey, M. D., Voglmayr, H., Brouwer, H., Bala, K., Chitty, D. W., Désaulniers, N., Eggertson, Q. A., Gachon, C. M. M., Hu, C. H., Küpper, F. C., Rintoul, T. L., Sarhan, E., Verstappen, E. C. P., Zhang, Y., Bonants, P. J. M., Ristaino, J. B., & Lévesque, C. A. (2011). DNAbarcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Molecular Ecology Resources, 11, 1002–1011.
Runge, F., Ndambi, B., Thines, M. (2012). Which morphological characteristics are most influenced by the host matrix in downy mildews? A case study in Pseudoperonospora cubensis. PLoS One, 7, e44863.
Savulescu, T. (1948). Les especes de Peronospora Corda de Roumanie. Sydowia, 2, 255–307.
Skalický, V. (1983). The revision of species of the genus Peronospora on host plants of the family Rosaceae with respect to Central European species. Folia Geobotanica et Phytotaxonomica, 18, 71–101.
Skalický, V. (1995). Agrimonia L.—řepík. In B. Slavík (Ed.), Květena České republiky 4 (Flora of the Czech Republic) (pp. 233–238). Praha: Academia (in Czech).
Smith, S., Roberson, S., & Cochran, K. (2014). First report of downy mildew on blackberry caused by Peronospora sparsa in Arkansas. Plant Disease, 98, 1585.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 4.0.. Molecular Biology and Evolution, 30, 2725–2729.
Thines, M., Telle, S., Ploch, S., & Runge, F. (2009). Identity of the downy mildew pathogens of basil, coleus, and sage with implications for quarantine measures. Mycological Research, 113, 532–540.
Tošovská, M., & Buchtová, I. (2012). Situační a výhledová zpráva léčivé, aromatické a kořeninové rostliny (Situation and Outlook Report Medicinal, Aromatic and Culinary Plants). Praha: Ministry of Agriculture (in Czech).
Voglmayr, H., Choi, Y. J., & Shin, H. D. (2014). Multigene phylogeny, taxonomy and reclassification of Hyaloperonospora on Cardamine. Mycological Progress, 13, 131–144.
Waller, J. M., Lenné, J. M., & Waller, S. J. (2001). Plant pathologist’s pocketbook. Wallingford: CABI Publishing.
White, T. J., Bruns, T., Lee, S., & Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. A. Innis, H. Gelfand, J. S. Sninsky, & T. J. White (Eds.), PCR protocols: A guide to methods and applications (pp. 315–322). San Diego: Academic.
Acknowledgments
Research was supported by: grant No. LO1204 (Sustainable development of research in the Centre of the Region Haná) from the National Program of Sustainability I, MEYS, Czech Republic; Internal Grant Agency of Palacký University in Olomouc, the Czech Republic (IGA 2016-001); fellowship from the Alexander von Humboldt foundation; LOEWE, Landesinitiative zur Entwicklung wissenschaftlich-ökonmischer Exzellenz, a funding program of the Government of Hessen, Germany, in the framework of the IPF—Integrative Fungal Research Cluster and the BiK-F—Biodiversity and Climate Research Centre. Authors thank to Prof. Aloisie Poulíčková for providing DIC microscopy photograph of P. agrimoniae conidiophore.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary file 1
(PDF 298 kb)
Rights and permissions
About this article
Cite this article
Petrželová, I., Choi, YJ., Jemelková, M. et al. Confirmation of Peronospora agrimoniae as a distinct species. Eur J Plant Pathol 147, 887–896 (2017). https://doi.org/10.1007/s10658-016-1058-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-016-1058-8