Abstract
Background & Aims
Kinesin family member 18A (KIF18A) is notable for its aberrant expression across various cancer types and its pivotal role is driving cancer progression. In this study, we aim to investigate the intricate molecular mechanisms underlying the impact of KIF18A on the progression of HCC.
Methods
Western blotting assays, a quantitative real-time PCR and immunohistochemical analyses were performed to quantitatively assess KIF18A expression in HCC tissues. We then performed genetic manipulations within HCC cells by silencing endogenous KIF18A using short hairpin RNA (shRNA) and introducing exogenous plasmids to overexpress KIF18A. We monitored cell progression, analyzed cell cycle and cell apoptosis and assessed cell migration and invasion both in vitro and in vivo. Moreover, we conducted RNA-sequencing to explore KIF18A-related signaling pathways utilizing Reactome and KEGG enrichment methods and validated these critical mediators in these pathways.
Results
Analysis of the TCGA-LIHC database revealed pronounced overexpression of KIF18A in HCC tissues, the finding was subsequently confirmed through the analysis of clinical samples obtained from HCC patients. Notably, silencing KIF18A in cells led to an obvious inhibition of cell proliferation, migration and invasion in vitro. Furthermore, in subcutaneous and orthotopic xenograft models, suppression of KIF18A sgnificantly redudce tumor weight and the number of lung metastatic nodules. Mechanistically, KIF18A appears to facilitate cell proliferation by upregulating MAD2 and CDK1/CyclinB1 expression levels, with the activation of SMAD2/3 signaling contributing to KIF18A-driven metastasis.
Conclusion
Our study elucidates the molecular mechanism by which KIF18A mediates proliferation and metastasis in HCC cells, offering new insights into potential therapeutic targets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC), the most common type of primary liver cancer, with the third leading cause of cancer-related mortalities in the world [1]. While innovative therapeutic strategies have been employed in the management of HCC, its progression remains marked by a poor prognosis, largely due to its high incidence of intrahepatic recurrence and extrahepatic metastasis [2, 3]. The gene aberrant expression accompanied with alterable progressions in the development of HCC multiple steps associated with increased cell proliferation, migration, and invasion [4, 5]. Consequently, an in-depth exploration of HCC’s pathogenesis and the pursuit of novel therapeutic targets to curb cancer cell metastasis represent a potential therapeutic strategy with the aim of enhancing the survival rates among HCC patients.
Kines family member-18A (KIF18A) is a member of the kinesin family comprising 45 members [6, 7]. Previous investigations have shown that KIF18A plays a pivotal role in maintaining kinetochore–microtubule connections and preserving centrosome integrity during mitosis [8]. These functions are crucial in the context of chromosomal instability (CIN) and the uncontrolled proliferation of tumor cells [8]. KIFA18A has also been associated with spindle alterations in aneuploid cells [9]. Furthermore, Kif18A’s sumoylation is a crucial factor in promoting successful mitotic progression [10]. Notably, KIF18A exhibits heightened expression in a wide spectrum of malignant tumors, including lung adenocarcinoma [11], prostate cancer [12], glioblastoma [13], and esophageal cancer [14], among others. In these malignancies, KIF18A proves to be indispensable for tumor cell proliferation, migration, and invasion. A study has suggested that KIF18A knockdown results in decreased migration of HCC cells, while the specific mechanism through which KIF18A influences HCC progression remains to be fully elucidated [15].
In this study, we found that KIF18A was abnormally highly expressed in clinical HCC tissues, and we validated that KIF18A knockdown suppressed HCC cell proliferation, migration, and invasion in vitro and in vivo firstly. Then, we demonstrated the underlying regulatory mechanisms driving the observed biological changes in HCC cells following KIF18A dysregulation; in detail, the KIF18A/MAD2/CDK1/CyclinB1 axis might be essential for HCC cell proliferation, while the activation of Smad2/3 signaling appears to be the key driver behind KIF18A-mediated metastasis. Consequently, our research positions KIF18A as a promising candidate for targeted interventions in the pathogenesis and metastatic progression of HCC.
Materials and Methods
Clinical Sample
Human HCC liver tissues and corresponding adjacent normal liver tissues (20 pairs) were collected and diagnosed by The First Affiliated Hospital of Chongqing Medical University, Chongqing, China. Written informed consent was obtained from each patient, and these scientific studies were approved by patients and the ethics committee.
Cell Culture
MHCC97-H, HLE, Huh-7, and PLC/PRF/5 cell lines were purchased form Guangzhou Jennio Biotech Co., Ltd., Guangzhou, China. All cells were validated by STR DNA fingerprinting and tested for mycoplasma. MHCC97-H and Huh-7 cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Sigma) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (pen/strep). HLE cells were cultured in minimum essential medium (MEM, Sigma) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. PLC/PRF/5 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Sigma) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. All cells were maintained at 37 °C in a humidified 5% CO2 atmosphere.
Establishment of KIF18A-Silenced Cells and Plasmids Transfection
The shRNAs targeting human KIF18A (sh-KIF18A#1, sh-KIF18A#2) and non-targeted control shRNA (shNC) were purchased from Shanghai GeneChem Co., Ltd, Shanghai, China. The shRNA targeting sequences were as follows: shKIF18A#1, 5′-TTGTTTCAGACTCACATATAA-3′; shKIF18A#2, 5′-AGTCCTGAGAGGAAGTCTTAA-3′; and shNC, 5′-TTCTCCGAACGTGTCACGT-3′. The MHCC97-H and HLE cell lines were infected with each shRNA-containing lentivirus and selected via puromycin (1 μg/mL). The establishment of stable cell lines was verified by Western blotting assay and quantitative real-time PCR assay of KIF18A. The KIF18A overexpression plasmid was constructed by subcloning its coding sequence into pcDNA3.1 empty vector utilizing BanHI and XhoI restriction enzymes. KIF18A plasmid or pcDNA3.1 empty vector was transfected into Huh-7 and PLC/PRF/5 cells using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. After 48 h, cells were harvested and verified by Western blotting assay and quantitative real-time PCR assay of KIF18A. All the verified cells were adopted for follow-up experiments.
Mice
Mouse model of subcutaneous tumor: 6-week-old male BALB/C nude mice were obtained from Beijing Huafukang Biotech (Beijing, China) and subcutaneously injected with KIF18A stable silenced MHCC97-H cells (3 × 106 cells in 100μl of PBS per mouse) in the thigh. The tumor-bearing mice were sacrificed by cervical dislocation. The tumor nodes were weighed and the tumor volume was calculated as: V (mm3) = width2 (mm2) × length (mm)/2.
Mouse model of orthotopic xenograft: 6-week-old male BALB/C nude mice were orthotopically injected with KIF18A stable silenced MHCC97-H cells (2 × 106 cells in 50 μL DMEM mixed with Matrigel as equal volume, per mouse) in the left hepatic lobe. The mice were sacrificed after 8 weeks of treatment, and liver tumor was harvested and photographed. The lung metastatic nodules and foci were counted though microscope observation after HE staining. All mouse studies were performed according to the protocol approved by the Chongqing Medical University Animal Care Committee and satisfied the “Guide for the Care and Use of Laboratory Animals” guidelines published by the US National Institutes of Health (NIH). All the mice were housed in a pathogen-free facility with a 12-h light, 12-h dark cycle and were provided with food and purified water ad libitum.
Quantitative Real-Time PCR
Total RNA was extracted from HCC cells with TRIzol reagent (Invitrogen, USA), and 1 μg RNA was reverse transcribed into cDNA using PrimerScript RT Kit (Bio-Rad, USA). Quantitative PCR was performed in three replicate wells on IQTM 5 Multicolor Real-Time PCR Detection system (Bio-Rad, USA) using FastStart Universal SYBR Green Master Mix (Roche, Basel, Switzerland). The target genes expression levels were normalized to β-actin RNA levels in each cell lines. The relative expression was calculated by the 2(−△△Ct) method. The primers are displayed in Supplementary Table 1.
Western Blotting Assay
Cell proteins were extracted using RIPA lysis buffer (#P0013, Beyotime, China) supplemented with 1% protease inhibitor cocktail tablets (#04693116001, Roche, Germany). Protein lysates concentration was quantified using BCA kits (#23225, Invitrogen, USA). The proteins were separated by SDS-PAGE on 8–12% gel and transferred to polyvinylidene difluoride (PVDF) membranes (GE Healthcare, Freiburg, Germany). The membranes were blocked with 5% non-fat milk and were incubated with primary antibody overnight at 4 °C. The primary antibodies included KIF18A antibody (#19245-1-AP, Proteintech, dilution 1:1000), CDK1 antibody (#19532-1-AP, Proteintech, dilution 1:1000), CyclinB1 antibody (#4138, CST, dilution 1:1000), MAD2 antibody (#10337-1-AP, Proteintech, dilution 1:1000), phospho-Smad2 antibody (#18338, CST, dilution 1:1000), phospho-Smad3 antibody (#9520, CST, dilution 1:1000), Smad2 antibody (#5339, CST, dilution 1:1000), Smad3 antibody (#66516-1-Ig, Proteintech, dilution 1:1000), E-cadherin antibody (#20874-1-AP, Proteintech, dilution 1:1000), N-cadherin antibody (#22018-1-AP, Proteintech, dilution 1:1000), α-Tubulin antibody (#2148, CST, dilution 1:1000), and GAPDH antibody (#MB001, Bioworld, dilution 1:1000). The membranes were incubated with secondary antibody before observing the targets with an ECL chemiluminescence system (e-BLOT Life Science, China) and densitometric analyzed with e-BLOT software. α-Tubulin and GAPDH were used as internal reference.
Flow Cytometry
Cell cycle and cell apoptosis were performed by flow cytometry. Annexin V—APC/propidium iodide (PI) apoptosis detection kit (#640932, BioLegend) was applied to detect the apoptosis according to the manufacturer’s manual. Briefly, cells were harvested and incubated with binding buffer supplemented with Annexin V and PI in dark for 15 min and analyzed using a FACScan flow cytometer (BD, Cytoflex, USA). For cell cycle detection, samples were harvested and fixed in 70% ethanol overnight at 4 °C, followed by treated with RNase (200 µg/mL) 30 min at 37 °C, subsequently incubated with PI (50 µg/mL) staining buffer for 15 min, and analyzed by flow cytometer. All samples were repeated three times.
Cell Counting Kit-8 Assay
Cells were plated in 96-well plates with the density at 1 × 104 cells/ml and incubated for 2 h, 12 h, 24 h, 36 h, 48 h, and 60 h. And then, each time point cell was incubated with CCK-8 solution (#96992, Sigma, USA) for 2 h at 37 °C. Multimode microplate reader (BioTek, USA) was performed to measure the absorbance at 450 nm of each well.
Wound Healing Assay
Cells were seeded to the 6-well plates and the monolayers cells were scratched with pipette tips at the confluence of 90%. The culture medium was supplemented with mitomycin C (10 mg/mL). The scratched area was imaged at 0 h and 48 h using an inverted microscope. Three locations of each image were chosen to calculate the migration rate (%) = (wound width 0 h − wound width at 48 h)/wound width 0 h × 100.
Transwell Assay
Cell migration was performed using a transwell insert (without Matrigel), and cell invasion was performed using pre-coated transwell insert (with 50 µL Matrigel). Cells were counted and resuspended in 500 µL serum-free DMEM medium before adding to the upper chamber, then the lower chamber was filled with DMEM containing 10% FBS. After 48 h of incubation, the reside cells on the upper chamber were removed and the cells under the membrane were fixed with methanol and stained with 1% crystal violet. Finally, the number of cells in five randomly selected fields was counted with inverted microscope. The experiment was repeated three times.
RNA-Sequencing Analysis
The RNA-sequencing analysis was performed by Shanghai Majorbio Bio-pharm Technology Co., Ltd (Shanghai, China). Each tested sample was pooled from three individual samples. The expression level of genes was analyzed in shKIF18A and shNC group by using DESeq2. P value < 0.05 and |log2FC| > 1.5 were considered significantly differentially expressed.
Immunohistochemistry
All the tissue sections were formalin-fixed and paraffin-embedded. After dewaxing and rehydration, the antigen of the slides was retrieved with 1 × EDTA antigen retrieval solution for 20 min at 95 °C. After quenching the endogenous peroxidase activity with H2O2 (3%), the slides were incubated with anti-KIF18A antibody overnight at 4 °C. Then, relative secondary antibody was added for incubation. The slides were visualized using diaminobenzidine (DAB) solution and then counterstained with hematoxylin. Immunoreactive score (IRS) system was performed to evaluate the immunohistochemical staining reaction.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism 8 (GraphPad Software, Inc.). Data were expressed as the means ± s.e.ms and were analyzed using a two-tailed unpaired Student’s t test. Paired t test was used to analyze the expression level in HCC tissues and corresponding adjacent normal liver tissues. P < 0.05 was considered statistically significant, for each parameter of all data presented, *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
KIF18A Expression Level Is Correlated with Clinicopathological Features in HCC Patients
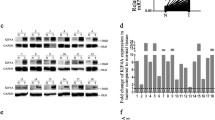
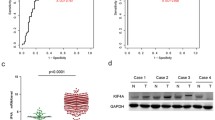
In this study, we undertook an extensive examination of the correlation between KIF18A mRNA expression levels and various clinical characteristics, utilizing data from The Cancer Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) database (http://ualcan.path.uab.edu/index.html). Our findings revealed a significant upregulation of KIF18A mRNA in primary HCC tumor tissues (n = 371) in stark contrast to the normal tissues (n = 50) (Fig. 1A). Furthermore, our analysis demonstrated that the expression of KIF18A increased with an increase in tumor grade and clinical stage (Fig. 1C, D); in detail, the highest KIF18A expression was found in grade 4 and stage 3, both indicated a highest degree of malignancy. High expression of KIF18A was correlated with poor disease-free survival and overall survival of patients with HCC (Fig. 1E, F). Interestingly, KIF18A mRNA expression did not show significant variation concerning nodal invasion (Fig. 1B). Additionally, we analyze the KIF18A mRNA expression in different tumor types from TCGA database which revealed that apart from kidney chromophobe (KICH), pancreatic adenocarcinoma (PAAD), pheochromocytoma and paraganglioma (PCPG), skin cutaneous melanoma (SKCM), and thymoma (THYM), KIF18A was highly expressed in nearly all the tumor tissues compared with normal tissues (Fig. 1G). In summary, our study indicates the pivotal role of aberrant KIF18A expression in relation to clinicopathological features, suggesting that its high expression may serve as a prognostic indicator for unfavorable outcomes.
The association between KIF18A expression and clinicopathological feature in TGCA database. A–D The KIF18A mRNA expression level in primary HCC tissues (n = 371) compared with normal tissues (n = 50) (B), and the KIF18A mRNA expression level between nodal invasion (B), and between tumor grade (C) and clinical stage (D). E and F Kaplan–Meler survival analysis of disease-free survival and overall survival associated with KIF18A expression in HCC patients. G The KIF18A expression level in pan-cancer tissues from the TGGA database. (***P < 0.001)
Expression of KIF18A in HCC Tissues and Adjacent Normal Liver Tissues
As mentioned previously, high expression of KIF18A in HCC tissues was found in the TCGA-LIHC database. In order to further corroborate this observation, we conducted a comprehensive investigation by examining KIF18A expression in 20 pairs of primary HCC and corresponding adjacent normal liver tissues. Then, we employed Western blotting assay to analyze the KIF18A protein expression in both HCC tissues and adjacent normal liver tissues. We found that protein level of KIF18A was significantly increased in HCCs compared with those adjacent normal liver tissues (Fig. 2A, B). Subsequently, we utilized quantitative real-time PCR to assess KIF18A mRNA level, and the results indicated that KIF18A mRNA expression level was elevated in HCCs (Fig. 2C). Furthermore, we conducted immunohistochemical staining assays to underscore our findings, and the results showed that KIF18A expression was higher in HCC tissues than in adjacent normal liver tissues (Fig. 2D, E). Further multivariate analysis found a positive correlation between KIF18A expression levels with tumor size, microvascular invasion, and metastasis, whereas no significant association was found between its expression levels and TNM stage (Fig. 2F). Conclusively, these results indicate that the expression of KIF18A in HCC tissues is significantly higher than in adjacent normal liver tissues, and high levels of KIF18A associate with aggressive malignant features in HCC patients.
The expression of KIF18A in HCC cancer tissues. A and B Western blotting assay detects the KIF18A protein expression levels in HCC tissues and corresponding normal liver tissues (n = 20 pairs) (A) and presents the gray value analysis (B). C Quantitative real-time PCR detects the KIF18A mRNA expression levels. D Immunohistochemical staining assays analyze the KIF18A expression in HCC tissues and corresponding normal liver tissues (n = 10 pairs). The magnification of the pictures from top to bottom is 200 times and 400 times. E The immunostaining tissues were scored by IRS. F Multivariant analysis of hazard ratios (HR) of KIF18A expression levels including tumor size, TNM stage, microvascular invasion, and metastasis. (***P < 0.001)
KIF18A Benefits the Proliferation of HCC Cells
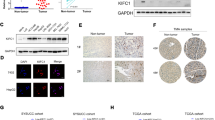
To determine the role of KIF18A in HCC cells, we established the stable knockdown of endogenous KIF18A in MHCC97-H and HLE cells, which have higher invasive and metastatic capabilities, by the utilization of lentivirus-mediated shRNAs. The efficiency of this interference was confirmed though quantitative real-time PCR (Fig. S1A) and Western blotting assay (Fig. S1C). KIF18A knockdown resulted in a considerable inhibitory effect on proliferation compared to shNC group as evidenced by CCK-8 assay (Fig. 3A, B). In contrast, ectopic expression of KIF18A exhibited the opposite effects in Huh-7 and PLC/PRF/5 cells, which have low metastatic potential (Fig. 3C, D). Moreover, KIF18A knockdown exhibited a notable increase in the G0/G1 and G2 proportions and a significant decrease in the S proportion in cell cycle progress, as contrasted with the shNC group, analyzed using flow cytometry (Fig. 3E), however, KIF18A-overexpressing decreased the proportions of cells in the G0/G1 and G2 phases and increase S proportions (Fig. 3F). Unexpectedly, neither knockdown nor overexpression of KIF18A affected cell apoptosis (Fig. S1D, E). We further examined the effect of KIF18A knockdown on tumor growth in subcutaneous xenograft model, and the results revealed that KIF18A knockdown markedly decreased the tumor size and volume (Fig. 3G, H).
Effect of KIF18A knockdown or overexpression on the proliferation of HCC cells. A and B Cell proliferation was analyzed by CCK-8 on KIF18A-silenced MHCC97-H and HLE. C and D Cell proliferation was analyzed by CCK-8 on KIF18A overexpression Huh-7 and PLC/PRF/5 cells. E Cell cycle distribution evaluated by using flow cytometry in KIF18A knockdown HCC cells. F Cell cycle distribution evaluated by using flow cytometry in KIF18A overexpression HCC cells. G and H The effect of KIF18A knockdown on tumor growth, tumor size, and tumor weight was evaluated by subcutaneous xenograft mouse model. I Reactome enrichment analysis enriched the differentially expressed genes after performing RNA-sequencing in KIF18A knockdown cells. J The effect of KIF18A knockdown or overexpression on CDK1, CyclinB1, and MAD2 proteins expression levels was tested by Western blotting assay. (**P < 0.01, ***P < 0.001)
To further investigate the underlying mechanism of KIF18A-promoting cell proliferation, we subjected KIF18A-silenced cells and their respective control to RNA-sequencing analysis. The differentially expressed genes were enriched through Reactome enrichment analysis and found that the cell cycle-related pathways including the packaging of telomere ends and meiotic synapsis were showed significant enrichment in KIF18A-silenced HCC cells (Fig. 3I). Telomeres, which represent the natural ends of linear chromosomes, comprise repeat-sequence DNA and associated proteins [16]. The replication of telomeres allows continued proliferation of human stem cells and immortality of cancer cells [16]. Intriguingly, MAD2 has been identified as a novel factor control mammalian telomers activity [17]. Previous reports have underscored the essential role of KIF18A and MAD2 in spindle assembly checkpoint-dependent mitotic arrest [8], and KIF18A knockdown induces mitotic delay in HPT cells [17], further implicating the intricate interplay between KIF18A and telomeric maintenance in cellular proliferation [18, 19]. Subsequently, we conducted an evaluation of mitotic-associated protein MAD2 and G2/M cell cycle-related proteins CDK1 and CyclinB1. The data of our Western blotting assay unequivocally demonstrated a notable reduction in the levels of MAD2, CDK1, and CyclinB1 within the shKIF18A group (Fig. 3J); in the meantime, the overexpression of KIF18A resulted in a significant increase in the expression of these proteins (Fig. 3J). Taken together, this finding collectively implies a functional linkage between the KIF18A/MAD2/CDK1 axis and the proliferation of HCC cells.
KIF18A Promote HCC Cell Migration and Invasion
We proceeded to investigate the role of KIF18A in HCC metastasis by preforming scratch wound assay and transwell assay. Based on scratch wound assay, KIF18A knockdown significantly diminished the wound healing capacity at 48 h in MHCC97-H and HLE cells (Fig. 4A, B), whereas the overexpression of KIF18A observed a noticeable reduction in cell scratch space in Huh-7 and PLC/PRF/5 cells at the same time point (Fig. 4C, D). Meanwhile, KIF18A knockdown markedly inhibited the migration and invasion capability of MHCC97-H and HLE cells (Fig. 4E, F), and the overexpression of KIF18A enhanced the migration and invasion abilities of Huh-7 and PLC/PRF/5 cells, as evidenced by transwell assay (Fig. 4G, H). Moreover, we injected the stable KIF18A knockdown MHCC97-H cells into the left lobe of orthotopic liver to establish a model of lung metastasis. The results revealed that KIF18A knockdown led to a significantly reduction in liver tumor volume (Fig. 4I) and the number of metastatic nodules and foci (Fig. 4J). Collectively, these in vitro and in vivo studies suggest that a pivotal role of KIF18A in promoting HCC metastasis.
Effect of KIF18A knockdown or overexpression on the migration and invasion of HCC cells. A and B The effect of KIF18A knockdown on cell migration was analyzed by wound healing assay in MHCC97-H and HLE cells. C and D The effect of KIF18A overexpression on cell migration was analyzed by wound healing assay in Huh-7 and PLC/PRF/5 cells. E and F The KIF18A knockdown on cell migration and invasion were analyzed by transwell assay in MHCC97-H and HLE cells. G and H The KIF18A overexpression on cell migration and invasion were analyzed by transwell assay in Huh-7 and PLC/PRF/5 cells. (I-J) Orthotopic xenograft mouse model was used to analyze the effect of KIF18A Knockdown on tumor growth and lung metastasis. (**P < 0.01, ***P < 0.001)
KIF18A Promotes HCC Cell Metastasis by Activating SMAD2/3 Signaling
Our subsequent investigation aimed to unravel the mechanism through which KIF18A promotes metastasis in HCC cells. The differentially expressed genes identified via RNA-seq were enriched by KEGG enrichment analysis. This analysis revealed a significant impact on the TGF-β signaling pathway in KIF18A-silenced HCC cell (Fig. 5A). In the canonical TGF-β signaling pathway, TGF-β receptor 2 phosphorylates TGF-β receptor 1 in response to TGF-β. The phosphorylated TGF-β receptor 1 subsequently phosphorylates SMAD2 and SMAD3. The phosphorylation of SMAD2/3 is the key step in initiating the TGF-β signaling pathway [20, 21]. The SMAD-dependent TGF-β signaling is known to promote metastasis by enhancing EMT and invasiveness in primary carcinomas [22,23,24,25]. Given the important role of the SMAD signaling in tumor metastasis, we then measured the phosphorylation levels of the key SMAD proteins and found that KIF18A silencing led to a decrease in the phosphorylation of SMAD2 and SMAD3 in MHCC97-H and HLE cells (Fig. 5B). Consistently, ectopic expression of KIF18A increased the phosphorylation levels of SMAD2 and SMAD3 in Huh-7 and PLC/PRF/5 cells (Fig. 5B). Notably, neither the silencing nor the overexpression of KIF18A had any discernible impact on the total SMAD2 and SMAD3 protein levels (Fig. 5B). It has been reported that the activated SMAD2/3 acts as a key transcription factor (TF) and interacts with other epithelial-to-mesenchymal transition (EMT)-associated TFs to regulate the expression of metastatic genes within the nuclear [26, 27].
The influence of KIF18A on TGF-β signaling pathway. A KEGG pathway enrichment analysis of the differentially expressed genes generated by RNA-seq showed that the TGF-β signaling pathway was significantly enriched, demonstrating the most statistically significant outcomes. B Western blotting assay was used to evaluate the effect of KIF18A knockdown or overexpression on p-SMA2/3, SMA2/3, E-cadherin, and N-cadherin proteins’ expression level in HCC cells. C Representative morphological images of KIF18A knockdown MHCC97-H cells and KIF18A-overexpresing Huh-7 cells. D and E Quantitative real-time PCR was used to analyze the effect of KIF18A knockdown or overexpression on EMT-associated genes’ expression level. F The possible signal pathway of KIF18A high expression affecting cell proliferation and cell metastasis in HCC cells. (*P < 0.05, **P < 0.01, ***P < 0.001)
Interestingly, EMT allows cells to acquire migration and invasion abilities during tumor metastasis, with the TGF-β signaling pathway serving as a key regulatory of EMT. We observed that in KIF18A knockdown MHCC97-H cells, the cell morphology changed from elongated mesenchymal form to an epithelial form. Conversely, in KIF18A overexpression Huh-7 cells, it shifted from an epithelial form to a spindle-shaped or elongated mesenchymal form (Fig. 5C), which indicated that KIF18A indeed functions to induce the EMT of HCC cell. Next, Western blotting assays were performed to ascertain the regulatory effect of KIF18A on epithelial (E-cadherin) and mesenchymal maker (N-cadherin) expressions. The results indicated that KIF18A knockdown remarkably enhanced E-cadherin expression but decreased N-cadherin expression, whereas the overexpression of KIF18A exhibited the opposite effects in Huh-7 cells (Fig. 5B). To identify the specific genes responsive to KIF18A, we then performed quantitative real-time PCR to screen SMAD2/3-regulated metastatic genes, including Snail1/2, ZEB1/2, Vim, and CDH1/2. The results revealed significant downregulation of Snail1/2, ZEB1, Vim, and CDH2 in KIF18A-silenced MHCC97-H and HLE cells (Fig. 5D). Conversely, these metastatic genes were upregulated in Huh-7 and PLC/PRF/5 cells upon overexpression of KIF18A (Fig. 5E). Together, these data demonstrate that KIF18A promotes HCC cell metastasis though the activation of SMAD2/3, leading to the upregulation of metastatic gene expression.
Discussion
HCC is a malignant tumor originating in the digestive tract, and its incidence has been increasing annually lately [1, 28]. Currently, surgical resection remains the primary treatment approach for patients with HCC, even though it is often combined with radiotherapy and chemotherapy, the 5-year survival rate of patients following surgical resection remains disappointingly low [2, 29, 30]. Therefore, it is of great urgency to investigate early diagnostic markers and potential therapeutic targets for this condition.
Kinesins are a family of molecular motor proteins that rely on microtubules [7]. The critical stages in the progression of cell mitosis involve the microtubule-associated spindle polymerization and the turnover of attachments between spindle microtubules and kinetochores [8, 31, 32]. KIF18A and MAD2 (a novel factor controls DNA repair activities at mammalian telomeres and essential for the spindle assembly checkpoint-dependent mitotic arrest), play a pivotal role in chromosome alignment in cells [8, 33,34,35]. Moreover, it promotes the satisfaction of the spindle assembly checkpoint and the progression through mitosis in certain cell types [9]. Knocking of KIF18A or MAD2 in cancer cells results in cell cycle arrest [8]. Several studies pointed out that KIF18A is upregulated in tumors and plays a critical role in tumor staging and patient prognosis [11, 12, 36] and KIF18A knockdown has the potential to suppress cell proliferation in many cancer types [6, 11, 13, 14, 37]. Furthermore, KIF18A has been proved to not only promote the proliferation of tumor cells, but also have the potential to promote the migration and invasion. In esophageal cancer, IGF2BP3 protein enhanced KIF18A mRNA stability to promote cell migration [14]. In colorectal cancer, KIF18A activates the PI3K/Akt signaling pathway by inhibiting PTEN transcription to promote migration and invasion in CRC cells [37]. In glioblastoma, KIF18A interacted with PPP1CA to promote GBM cell migration and invasion [38]. However, the exact role of KIF18A in HCC proliferation and migration has not been comprehensively elucidated as of yet [15].
In this study, we further explored the mechanism of KIF18A in regulating HCC. Firstly, we observed that the expression of KIF18A in HCC tissues was higher than adjacent normal liver tissues. Subsequently, upon silencing KIF18A in MHCC97-H and HLE cell lines, we noted a substantial reduction in cell proliferation, migration, and invasion. Additionally, in a tumor-bearing mouse model, the shKIF18A group exhibited significantly reduced tumor volume and fewer lung metastatic nodes. For the mechanism of KIF18A-promoting HCC proliferation, we detected the expression of some proteins based on the previous reports, namely MAD2, CDK1, and CyclinB1. MAD2 required for mitotic spindle [39, 40], CDK1 acts as the communicator in cell division, and CyclinB1 regulates mitosis in the G2/M phase of the cell cycle [41,42,43,44]. Our results validated that elevated KIF18A could promote the expression of MAD2 and CDK1/CyclinB1 proteins, which are necessary for cell proliferation. While our study focused solely on protein expression, the detailed molecular mechanism between KIF18A and MAD2/CDK1/CyclinB1 axis requires further in-depth research and confirmation.
For the mechanism of KIF18A promote HCC metastasis, we employed KEGG pathway analysis following RNA-sequencing in KIF18A-silenced cells. This analysis revealed significant alterations in the TGF-β signaling pathway, a critical player in cancer metastasis [20]. The activated TGF-β1 phosphorylates SMAD2/SMAD3, leading to their translocation into the nucleus for transcriptional regulation, which, in turn, induces the expression of vimentin and N-cadherin, thereby promoting epithelial–mesenchymal transition (EMT) and cancer metastasis [26, 45]. Inhibition of the TGF-β-SMAD2/3 pathway has been found to suppress cancer metastasis [45, 46]. Moreover, phosphorylated SMAD2/SMAD3 targets transcription factors, including SNAIL, ZEB1/2, c-Myc, C/EBPβ, and others. SNAIL1/2 and ZEB1/2 are the principal transcription factors involved in regulating EMT progression [26]. Here, our findings demonstrated that KIF18A significantly elevated phosphorylated SMAD2/SMAD3 proteins levels and upregulated the expression of SNAIL1/2, ZEB1, and their downstream EMT-associated target genes.
Now, we know that the high expression of KIF18A plays an important role in many caners, including but not limited to colon [37], breast [6], esophageal [14], prostate [12], and glioblastoma [13]. KIF18A appears to be a viable target for the treatment of cancer and a series of inhibitors is identified [47]. BTB1, the first identified KIF18A inhibitor, and its analogs were observed to have potent ability to inhibit the ATPase activity of KIF18A [48, 49], however, which have a great impact on highly selective and bioactive needed further investigation [50]. At present, the application of those inhibitors is still in the stage of cell experiments, more detailed evidence is needed for prospective clinical application. The poor prognosis of HCC remains a challenge due to high rates of metastasis and postoperative recurrence [51, 52]. KIF18A is frequently overexpression in HCC, the therapeutic strategies targeting KIF18A are not only of great interest for basic research but also have the potential to the new strategies for the treatment of patients with multiple metastases or prevention the recurrence after resection.
In conclusion, the findings of this study highlight the potential role of the KIF18A/MAD2 axis in promoting cell proliferation, and the KIF18A-SMAD2/3 signaling pathway appears to be accountable for cell migration and invasion in HCC cells. Therefore, KIF18A represents a promising target for the development of mechanism-based HCC prevention strategies.
Data availability
Additional supporting data are available from the corresponding authors on request. All requests for raw and analyzed data and materials will be reviewed by the corresponding authors to verify whether the request is subject to any intellectual property or confidentiality obligations.
References
Sung H, Ferlay J, Siegel RL et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA 2021;71:209–249.
Llovet JM, Kelley RK, Villanueva A et al. Hepatocellular carcinoma. Nat Rev. 2021;7:6.
Yang G, Xiong Y, Sun J et al. The efficacy of microwave ablation versus liver resection in the treatment of hepatocellular carcinoma and liver metastases: a systematic review and meta-analysis. Int J Surg. 2020;77:85–93.
Yang B, Li M, Tang W et al. Dynamic network biomarker indicates pulmonary metastasis at the tipping point of hepatocellular carcinoma. Nat Commun 2018;9:1. https://doi.org/10.1038/s41467-018-03024-2.
Qi F, Li J, Qi Z et al. Comprehensive metabolic profiling and genome-wide analysis reveal therapeutic modalities for hepatocellular carcinoma. Research. 2023. https://doi.org/10.34133/research.0036.
Alfarsi LH, Elansari R, Toss MS et al. Kinesin family member-18A (KIF18A) is a predictive biomarker of poor benefit from endocrine therapy in early ER+ breast cancer. Breast Cancer Res Treat 2019;173:93–102.
Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell 1985;42:39–50.
Marquis C, Fonseca CL, Queen KA et al. Chromosomally unstable tumor cells specifically require KIF18A for proliferation. Nat Commun. 2021. https://doi.org/10.1038/s41467-021-21447-2.
Janssen LME, Averink TV, Blomen VA et al. Loss of Kif18A results in spindle assembly checkpoint activation at microtubule-attached kinetochores. Current Biol 2018;28:2685–2696.
Yang F, Chen Y, Dai W. Sumoylation of Kif18A plays a role in regulating mitotic progression. BMC Cancer. 2015. https://doi.org/10.1186/s12885-015-1226-9.
Zhong Y, Jiang L, Lin H et al. Overexpression of KIF18A promotes cell proliferation, inhibits apoptosis, and independently predicts unfavorable prognosis in lung adenocarcinoma. IUBMB Life 2019;71:942–955.
Zhang H, Shen T, Zhang Z et al. Expression of KIF18A Is associated with increased tumor stage and cell proliferation in prostate cancer. Med Sci Monit. 2019;25:6418–6428.
Wang L, Zhang X, Liu J et al. The proliferation of glioblastoma is contributed to kinesin family member 18A and medical data analysis of GBM. Front Genet. 2022. https://doi.org/10.3389/fgene.2022.858882.
Qian L, Cao X, Du M et al. KIF18A knockdown reduces proliferation, migration, invasion and enhances radiosensitivity of esophageal cancer. Biochem Biophys Res Commun. 2021;557:192–198.
Luo W, Liao M, Liao Y et al. The role of kinesin KIF18A in the invasion and metastasis of hepatocellular carcinoma. World J Surg Oncol. 2018. https://doi.org/10.1186/s12957-018-1342-5.
Lim CJ, Cech TR. Shaping human telomeres: from shelterin and CST complexes to telomeric chromatin organization. Nat Rev. 2021;4:283–298.
Boersma V, Moatti N, Segura-Bayona S et al. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5′ end resection. Nature 2015;521:537–540.
Meadows JC, Lancaster TC, Buttrick GJ et al. Identification of a Sgo2-dependent but Mad2-independent pathway controlling anaphase onset in fission yeast. Cell Rep. 2017;18:1422–1433.
Lee SH, McCormick F, Saya H. Mad2 inhibits the mitotic kinesin MKlp2. J Cell Biol. 2010;191:1069–1077.
Massagué J. TGFβ in cancer. Cell 2008;2:215–230.
Gough NR, Xiang X, Mishra L. TGF-beta signaling in liver, pancreas, and gastrointestinal diseases and cancer. Gastroenterology 2021;161:434–452.
Wu MZ, Yuan YC, Huang BY et al. Identification of a TGF-β/SMAD/lnc-UTGF positive feedback loop and its role in hepatoma metastasis. Signal Transd Targeted Therapy 2021;395:1–14.
Zheng L, Liang H, Zhang Q et al. circPTEN1, a circular RNA generated from PTEN, suppresses cancer progression through inhibition of TGF-β/Smad signaling. Mol Cancer. 2022. https://doi.org/10.1186/s12943-022-01495-y.
Tuncer E, Calçada RR, Zingg D et al. SMAD signaling promotes melanoma metastasis independently of phenotype switching. J Clin Investig 2019;129:2702–2716.
Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial–mesenchymal-like transition and promotes metastasis. Cancer Cell 2011;20:576–590.
Yeh H, Hsu E, Lee S et al. PSPC1 mediates TGF-β1 autocrine signalling and Smad2/3 target switching to promote EMT, stemness and metastasis. Nat Cell Biol 2018;20:479–491.
Zhang L, Zhu Z, Yan H et al. Creatine promotes cancer metastasis through activation of Smad2/3. Cell Metab 2021;33:1111–1123.
Siegel RL, Miller KD, Fuchs HE et al. Cancer statistics, 2021. CA 2021;71:7–33.
Wang Z, Ren Z, Chen Y et al. Adjuvant transarterial chemoembolization for HBV-related hepatocellular carcinoma after resection: a randomized controlled study. Clin Cancer Res. 2018;24:2074–2081.
Ricke J, Klümpen HJ, Amthauer H et al. Impact of combined selective internal radiation therapy and sorafenib on survival in advanced hepatocellular carcinoma. J Hepatol. 2019;71:1164–1174.
Ertych N, Stolz A, Stenzinger A et al. Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat Cell Biol. 2014;16:779–791.
Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Current Biol. 2009;19:1937–1942.
Czechanski A, Kim H, Byers C et al. Kif18a is specifically required for mitotic progression during germ line development. Dev Biol. 2015;402:253–262.
Fonseca CL, Malaby HLH, Sepaniac LA et al. Mitotic chromosome alignment ensures mitotic fidelity by promoting interchromosomal compaction during anaphase. J Cell Biol. 2019;218:1148–1163.
Stumpff J, von Dassow G, Wagenbach M et al. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell 2008;14:252–262.
Tao B, Liu Y, Liu H et al. Prognostic biomarker KIF18A and its correlations with immune infiltrates and mitosis in glioma. Front Genet. 2022. https://doi.org/10.3389/fgene.2022.852049.
Liu Y, Sun M, Zhang B et al. KIF18A improves migration and invasion of colorectal cancer (CRC) cells through inhibiting PTEN signaling. Aging (Albany NY) 2023;15:9182–9192.
Yang J, Zhang Q, Yang Z et al. KIF18A interacts with PPP1CA to promote the malignant development of glioblastoma. Exp Ther Med. 2023;25:154.
Luo X, Tang Z, Xia G et al. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11:338–345.
Shimada K, Gasser SM. The origin recognition complex functions in sister-chromatid cohesion in Saccharomyces cerevisiae. Cell 2007;128:85–99.
Yu J, Raia P, Ghent CM et al. Structural basis of human separase regulation by securin and CDK1–cyclin B1. Nature 2021;596:138–142.
Chen N, Aretz J, Fässler R. CDK1–cyclin-B1-induced kindlin degradation drives focal adhesion disassembly at mitotic entry. Nat Cell Biol. 2022;24:723–736.
Wang Z, Fan M, Candas D et al. Cyclin B1/Cdk1 coordinates mitochondrial respiration for cell-cycle G2/M progression. Dev Cell 2014;29:217–232.
Clemm Von Hohenberg K, Müller S, Schleich S et al. Cyclin B/CDK1 and Cyclin A/CDK2 phosphorylate DENR to promote mitotic protein translation and faithful cell division. Nat Commun. 2022. https://doi.org/10.1038/s41467-022-28265-0.
Calon A, Espinet E, Palomo-Ponce S et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cell 2012;22:517–584.
Lee JH, Massagué J. TGF-β in developmental and fibrogenic EMTs. Semin Cancer Biol 2022;86:136–145.
Braun J, Mockel MM, Strittmatter T et al. Synthesis and biological evaluation of optimized inhibitors of the mitotic kinesin Kif18A. ACS Chem Biol 2015;10:554–560.
Catarinella M, Grüner T, Strittmatter T et al. BTB-1: a small molecule inhibitor of the mitotic motor protein Kif18A. Angew Chem Int Ed. 2009;48:9072–9076.
Sabnis RW. Novel KIF18A inhibitors for treating cancer. ACS Med Chem Lett. 2020;11:2368–2369.
Sabnis RW. Novel amide compounds as KIF18A inhibitors for treating cancer. ACS Med Chem Lett. 2021;12:690–691.
Ryu SH, Jang MK, Kim WJ et al. Metastatic tumor antigen in hepatocellular carcinoma: golden roads toward personalized medicine. Cancer Metast Rev 2014;33:965–980.
Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell 2017;168:670–691.
Funding
This work was supported by Chongqing Natural Science Foundation (Grant No. CSTB2022NSCQ-MSX0864) and Science and Technology Research Project of Chongqing Municipal Education Commission (Grant No. KJQN202100429).
Author information
Authors and Affiliations
Contributions
DPZ and JHR designed the study. XYY, MLY, STC, and DQW performed the experiments. KXX, RRL, and HZ analyzed the data. DPZ wrote the manuscript. JHR reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that there are no potential conflicts of interest.
Ethical approval
The studies involving human participants were reviewed and approved by The Research Ethics Committee of the First Affiliated Hospital of Chongqing Medical University in Southwest China. The patients/participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the guidelines of Chongqing Medical University Animal Care Committee.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Ren, J., Yao, X., Yang, M. et al. Kinesin Family Member-18A (KIF18A) Promotes Cell Proliferation and Metastasis in Hepatocellular Carcinoma. Dig Dis Sci 69, 1274–1286 (2024). https://doi.org/10.1007/s10620-024-08321-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-024-08321-z