Abstract
Background
Gallbladder cancer (GBC) remains a serious cause of cancer-related mortality across the globe. E2F5 has been identified to as a known oncogene in various cancers. However, the special functions of E2F5 have not been investigated in GBC.
Aims
To explore the regulatory functions of E2F5 and its related molecular regulatory mechanism in GBC progression.
Methods
The expression of genes were examined through qRT-PCR, western blot and IHC assay. The cell proliferation was assessed through CCK-8 and EDU assays. The cytotoxicity was tested through LDH assay. The percentage of CD8+ T cells and cell apoptosis were evaluated through flow cytometry. The binding ability was detected through luciferase reporter assay. The tumor growth was assessed through in vivo assays.
Results
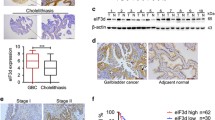
In this study, it was demonstrated that E2F5 expression was evaluated in GBC, and resulted into poor prognosis. Bioinformatics analysis revealed E2F5 as a target for let-7d-5p, which when overexpressed, suppressed the metastasis and proliferation of GBC through the downregulation of E2F5. It was discovered that E2F5 activates JAK2/STAT3 signaling which is suppressed by let-7d-5p, implicating this pathway as one of the effectors of the oncogenic effects of ESF5 in GBC. E2F5 had been confirmed to aggravate tumor growth in vivo.
Conclusion
E2F5 targeted by let-7d-5p facilitated cell proliferation, metastasis and immune escape in GBC through the JAK2/STAT3 pathway.
Graphical Abstract







Similar content being viewed by others
References
Misra S, Chaturvedi A, Misra NC, Sharma ID. Carcinoma of the gallbladder. Lancet Oncol 2003;4:167–176.
Andia ME, Hsing AW, Andreotti G, Ferreccio C. Geographic variation of gallbladder cancer mortality and risk factors in Chile: a population-based ecologic study. Int J Cancer 2008;123:1411–1416.
Ertel AE, Bentrem D, Abbott DE. Gall bladder cancer. Cancer Treat Res 2016;168:101–120.
Schmidt MA, Marcano-Bonilla L, Roberts LR. Gallbladder cancer: epidemiology and genetic risk associations. Chin Clin Oncol 2019;8:31.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424.
Hundal R, Shaffer EA. Gallbladder cancer: epidemiology and outcome. Clin Epidemiol 2014;6:99–109.
Pilgrim CH, Groeschl RT, Christians KK, Gamblin TC. Modern perspectives on factors predisposing to the development of gallbladder cancer. HPB (Oxford) 2013;15:839–844.
Iyer P, Barreto SG, Sahoo B, Chandrani P, Ramadwar MR, Shrikhande SV et al. Non-typhoidal Salmonella DNA traces in gallbladder cancer. Infect Agent Cancer 2016;11:12.
Sasatomi E, Tokunaga O, Miyazaki K. Precancerous conditions of gallbladder carcinoma: overview of histopathologic characteristics and molecular genetic findings. J Hepatobiliary Pancreat Surg 2000;7:556–567.
Wistuba II, Tang M, Maitra A, Alvarez H, Troncoso P, Pimentel F et al. Genome-wide allelotyping analysis reveals multiple sites of allelic loss in gallbladder carcinoma. Cancer Res 2001;61:3795–3800.
Rashid A. Cellular and molecular biology of biliary tract cancers. Surg Oncol Clin N Am 2002;11:995–1009.
Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 2002;3:11–20.
Attwooll C, Lazzerini Denchi E, Helin K. The E2F family: specific functions and overlapping interests. Embo j 2004;23:4709–4716.
Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer 2009;9:785–797.
Xie H, Kang Y, Wang S, Zheng P, Chen Z, Roy S et al. E2f5 is a versatile transcriptional activator required for spermatogenesis and multiciliated cell differentiation in zebrafish. PLoS Genet 2020;16:e1008655.
Ishimoto T, Shiozaki A, Ichikawa D, Fujiwara H, Konishi H, Komatsu S et al. E2F5 as an independent prognostic factor in esophageal squamous cell carcinoma. Anticancer Res 2013;33:5415–5420.
Li L, Liu J, Huang W. E2F5 promotes proliferation and invasion of gastric cancer through directly upregulating UBE2T transcription. Dig Liver Dis 2022;54:937–945.
Malgundkar SH, Burney I, Al Moundhri M, Al Kalbani M, Lakhtakia R, Okamoto A et al. E2F5 promotes the malignancy of ovarian cancer via the regulation of hippo and Wnt pathways. Genet Test Mol Biomark 2021;25:179–186.
Inagaki Y, Wu D, Fujiwara K, Ishizuka Y, Oguni A, Tokunaga T et al. Knockdown of E2F5 induces cell death via the TP53-dependent pathway in breast cancer cells carrying wild-type TP53. Oncol Rep 2020;44:2241–2252.
Karmakar D, Maity J, Mondal P, Shyam Chowdhury P, Sikdar N, Karmakar P et al. E2F5 promotes prostate cancer cell migration and invasion through regulation of TFPI2, MMP-2 and MMP-9. Carcinogenesis 2020;41:1767–1780.
Abrahamsson A, Dabrosin C. Tissue specific expression of extracellular microRNA in human breast cancers and normal human breast tissue in vivo. Oncotarget 2015;6:22959–22969.
Poursadegh Zonouzi AA, Nejatizadeh A, Rahmati-Yamchi M, Fardmanesh H, Shakerizadeh S, Poursadegh Zonouzi A et al. Dysregulated expression of Dicer in invasive ductal breast carcinoma. Med Oncol 2015;32:203.
Long J, Ou C, Xia H, Zhu Y, Liu D. MiR-503 inhibited cell proliferation of human breast cancer cells by suppressing CCND1 expression. Tumour Biol 2015;36:8697–8702.
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–233.
Martignetti L, Tesson B, Almeida A, Zinovyev A, Tucker GC, Dubois T et al. Detection of miRNA regulatory effect on triple negative breast cancer transcriptome. BMC Genomics 2015;16:S4.
Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 2010;11:252–263.
Yang MH, Yu J, Jiang DM, Li WL, Wang S, Ding YQ. microRNA-182 targets special AT-rich sequence-binding protein 2 to promote colorectal cancer proliferation and metastasis. J Transl Med 2014;12:109.
Chandra V, Kim JJ, Mittal B, Rai R. MicroRNA aberrations: An emerging field for gallbladder cancer management. World J Gastroenterol 2016;22:1787–1799.
Zhao M, Ang L, Huang J, Wang J. MicroRNAs regulate the epithelial-mesenchymal transition and influence breast cancer invasion and metastasis. Tumour Biol 2017;39:1010428317691682.
Li S, Liu F, Xu L, Li C, Yang X, Guo B et al. Wnt/β-catenin signaling axis is required for TFEB-mediated gastric cancer metastasis and epithelial-mesenchymal transition. Mol Cancer Res 2020;18:1650–1659.
Sun HD. MicroRNA-195-5p inhibits malignant progression of gallbladder cancer by regulating Wnt3a. Eur Rev Med Pharmacol Sci 2021;25:7635–7642.
Choo KB, Soon YL, Nguyen PN, Hiew MS, Huang CJ. MicroRNA-5p and -3p co-expression and cross-targeting in colon cancer cells. J Biomed Sci 2014;21:95.
Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle 2009;8:843–852.
Koh W, Sheng CT, Tan B, Lee QY, Kuznetsov V, Kiang LS et al. Analysis of deep sequencing microRNA expression profile from human embryonic stem cells derived mesenchymal stem cells reveals possible role of let-7 microRNA family in downstream targeting of hepatic nuclear factor 4 alpha. BMC Genomics 2010;11:S6.
Coskun M, Bjerrum JT, Seidelin JB, Troelsen JT, Olsen J, Nielsen OH. miR-20b, miR-98, miR-125b-1*, and let-7e* as new potential diagnostic biomarkers in ulcerative colitis. World J Gastroenterol 2013;19:4289–4299.
Wu L, Zhao Q, Zhu X, Peng M, Jia C, Wu W et al. A novel function of microRNA let-7d in regulation of galectin-3 expression in attention deficit hyperactivity disorder rat brain. Brain Pathol 2010;20:1042–1054.
Koh YW, Jung SJ, Park CS, Yoon DH, Suh C, Huh J. LGALS3 as a prognostic factor for classical Hodgkin’s lymphoma. Mod Pathol 2014;27:1338–1344.
Jain K, Sreenivas V, Velpandian T, Kapil U, Garg PK. Risk factors for gallbladder cancer: a case-control study. Int J Cancer 2013;132:1660–1666.
Polanowska J, Le Cam L, Orsetti B, Vallés H, Fabbrizio E, Fajas L et al. Human E2F5 gene is oncogenic in primary rodent cells and is amplified in human breast tumors. Genes Chromosomes Cancer 2000;28:126–130.
Tian H, Hou L, Xiong YM, Huang JX, Zhang WH, Pan YY et al. miR-132 targeting E2F5 suppresses cell proliferation, invasion, migration in ovarian cancer cells. Am J Transl Res 2016;8:1492–1501.
Xu H, Fei D, Zong S, Fan Z. MicroRNA-154 inhibits growth and invasion of breast cancer cells through targeting E2F5. Am J Transl Res 2016;8:2620–2630.
Lin C, Hu Z, Yuan G, Su H, Zeng Y, Guo Z et al. MicroRNA-1179 inhibits the proliferation, migration and invasion of human pancreatic cancer cells by targeting E2F5. Chem Biol Interact 2018;291:65–71.
Barh D, Malhotra R, Ravi B, Sindhurani P. MicroRNA let-7: an emerging next-generation cancer therapeutic. Curr Oncol 2010;17:70–80.
Tsunekuni K, Konno M, Asai A, Koseki J, Kobunai T, Takechi T et al. MicroRNA profiles involved in trifluridine resistance. Oncotarget 2017;8:53017–53027.
Fu LX, Lian QW, Pan JD, Xu ZL, Zhou TM, Ye B. JAK2 tyrosine kinase inhibitor AG490 suppresses cell growth and invasion of gallbladder cancer cells via inhibition of JAK2/STAT3 signaling. J Biol Regul Homeost Agents 2017;31:51–58.
Liu Y, Bi T, Yuan F, Gao X, Jia G, Tian Z. S-adenosylmethionine induces apoptosis and cycle arrest of gallbladder carcinoma cells by suppression of JAK2/STAT3 pathways. Naunyn Schmiedebergs Arch Pharmacol 2020;393:2507–2515.
Wang H, Zhan M, Liu Q, Wang J. Glycochenodeoxycholate promotes the metastasis of gallbladder cancer cells by inducing epithelial to mesenchymal transition via activation of SOCS3/JAK2/STAT3 signaling pathway. J Cell Physiol 2020;235:1615–1623.
Funding
This work was supported by the China State Railway Group Co., Ltd. (J2022Z615).
Author information
Authors and Affiliations
Contributions
LC, SYG wrote the manuscript, DFZ, XYL, JFC collected and analyzed the data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Fig.S1 Knockdown or overexpression of E2F5 has no effects on the other members of the E2F family. A
The protein expressions of E2F1, E2F2, E2F6, E2F8 were examined after silencing E2F5 through WB. The other isoforms of E2F were not affected through downregulation via siRNA. B The protein expressions of E2F1, E2F2, E2F6, E2F8 were tested after overexpressing E2F5 through WB. The other isoforms of E2F were not affected after E2F5 overexpression. All the data are from three independent experiments (JPG 1854 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, L., Guo, S., Zhang, D. et al. E2F5 Targeted by Let-7d-5p Facilitates Cell Proliferation, Metastasis and Immune Escape in Gallbladder Cancer. Dig Dis Sci 69, 463–475 (2024). https://doi.org/10.1007/s10620-023-08209-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-023-08209-4