Abstract
Background
We recently demonstrated that diarrhea-predominant irritable bowel syndrome (IBS-D) subjects have higher relative abundance (RA) of hydrogen sulfide (H2S)-producing Fusobacterium and Desulfovibrio species, and constipation-predominant IBS (IBS-C) subjects have higher RA of methanogen Methanobrevibacter smithii.
Aims
In this study, we investigate the effects of increased methanogens or H2S producers on stool phenotypes in rat models.
Methods
Adult Sprague–Dawley rats were fed high-fat diet (HFD) for 60 days to increase M. smithii levels, then gavaged for 10 days with water (controls) or methanogenesis inhibitors. To increase H2S producers, rats were gavaged with F. varium or D. piger. Stool consistency (stool wet weight (SWW)) and gas production were measured. 16S rRNA gene sequencing was performed on stool samples.
Results
In HFD diet-fed rats (N = 30), stool M. smithii levels were increased (P < 0.001) after 52 days, correlating with significantly decreased SWW (P < 0.0001) at 59 days (R = − 0.38, P = 0.037). Small bowel M. smithii levels decreased significantly in lovastatin lactone-treated rats (P < 0.0006), and SWW increased (normalized) in lovastatin hydroxyacid-treated rats (P = 0.0246), vs. controls (N = 10/group). SWW increased significantly in D. piger-gavaged rats (N = 16) on day 10 (P < 0.0001), and in F. varium-gavaged rats (N = 16) at all timepoints, vs. controls, with increased stool H2S production. 16S sequencing revealed stool microbiota alterations in rats gavaged with H2S producers, with higher relative abundance (RA) of other H2S producers, particularly Lachnospiraceae and Bilophila in F. varium-gavaged rats, and Sutterella in D. piger-gavaged rats.
Conclusions
These findings suggest that increased M. smithii levels result in a constipation-like phenotype in a rat model that is partly reversible with methanogenesis inhibitors, whereas gavage with H2S producers D. piger or F. varium results in increased colonization with other H2S producers and diarrhea-like phenotypes. This supports roles for the increased RA of methanogens and H2S producers identified in IBS-C and IBS-D subjects, respectively, in contributing to stool phenotypes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Irritable bowel syndrome (IBS) is a multifactorial gastrointestinal disorder that affects approximately one in ten people worldwide, although prevalence estimates vary widely [1]. Characteristic symptoms include abdominal pain, bloating, and changes in stool form and frequency that begin at least 6 months prior to diagnosis [2]. The three main subtypes are diarrhea-predominant IBS (IBS-D), constipation-predominant IBS (IBS-C), and mixed IBS with constipation and diarrhea (IBS-M) [2].
There is strong evidence that the gut microbiome plays an important role in the pathophysiology of IBS [3]. A meta-analysis of 23 case–control studies identified some consistent findings when stool samples from IBS subjects were compared to those from healthy controls, including a decrease in fecal levels of the genera Lactobacillus and Bifidobacterium in IBS subjects, as well as increases in fecal levels of family Enterobacteriaceae and species Escherichia coli [4]. A smaller earlier meta-analysis also identified lower Lactobacillus and Bifidobacterium levels in IBS subjects, as well as lower Faecalibacterium prausnitzii, when compared to healthy controls [5]. However, there is increasing evidence that IBS subtypes should be studied separately, as IBS-C and IBS-D appear to be associated with different gut microbial compositions (microtypes) [6]. Specifically, subjects with IBS-C have higher levels of methane (CH4)-producing archaea in their stool when compared to healthy controls [7], including higher levels of Methanobrevibacter smithii [8], which is the predominant methanogen in the human gut. Moreover, levels of this methanogen correlate with CH4 levels on a lactulose breath test [8,9,10]. In a recent publication, we identified distinct gut microbial compositions and associated breath gas profiles in subjects with IBS-C vs. IBS-D, with increased breath CH4 in IBS-C subjects that correlated with increased levels of methanogens, including M. smithii, in stool samples, whereas IBS-D subjects had increased breath levels of both hydrogen (H2) and hydrogen sulfide (H2S) [6]. A recent study by Algera et al., found that increased breath CH4 levels were associated with decreased oroanal transit times in IBS subjects, and while that study did not measure H2S levels, they did find associations between increased breath H2 levels and increased oroanal transit times in IBS [11]. The authors proposed that these findings resulted from differences in gut microbial compositions in different groups of IBS subjects [11], which is consistent with our identification of distinct gut microtypes and breath gas profiles in IBS-C vs. IBS-D subjects [6]. Further, we found that increased breath H2S levels in IBS-D subjects correlated with increased abundances of H2S-producing bacterial species, including species from the genera Fusobacterium and Desulfovibrio, in stool samples [6], and microbial metabolic pathway analysis suggested enrichment of KEGG modules associated with methane production in CH4-positive IBS-C subjects, and enrichment of modules associated with H2S production in IBS-D subjects [6]. While these findings suggest that increased M. smithii and H2S producers may contribute to stool phenotypes in IBS-C and IBS-D subjects, respectively, these are associations and do not prove cause-and-effect. In this study, we used rat models to explore the effects of increased levels of methanogens or H2S producers on stool phenotypes, in order to identify the potential microbial drivers of phenotypes seen in the different IBS subtypes.
Methods
Adult male Sprague Dawley rats (Envigo, Madison, WI) were used to test the effects of (1) the methanogen M. smithii, and (2) the H2S producers Desulfovibrio piger and Fusobacterium varium, on stool phenotypes. Rats were housed under regular light–dark cycles of 12/12 h and had free access to food and water at all times. Both studies were approved by the Cedars-Sinai Institutional Animal Care and Use Committee (IACUC).
Effects of Methanobrevibacter smithii on Stool Consistency
Previous work showed that Sprague–Dawley rats are endogenously colonized with M. smithii and that M. smithii gavage only transiently augmented its absolute abundance [12]. However, when rats were placed on high-fat diet (HFD), M. smithii levels increased significantly and remained stable for the duration of HFD feeding [12]. Therefore, HFD was used to assess the effects of persistent M. smithii elevation on stool consistency (% wet weights). Sprague–Dawley rats were placed on a HFD (60% energy from fat [D12492; Research Diets, New Brunswick, NJ]). Three-day stool collections (for determination of % wet weights [13]) were performed at baseline and on days 12, 59, and 71, and M. smithii levels were determined at baseline, on days 3 and 52, and at euthanasia (Fig. 1). To confirm that any effects on stool consistency were associated with absolute levels of M. smithii, and not secondary HFD effects, rats were subsequently divided into 3 groups and gavaged for 10 days with either 1.5 mg/ml lovastatin lactone, 1.5 mg/ml lovastatin hydroxyacid, or water (as controls), after which additional stool collections were performed (Fig. 1). Lovastatin is a natural inhibitor of methanogens, with the lactone and hydroxyacid forms having different mechanisms of action. Specifically, lovastatin hydroxyacid can inhibit archaeal cell wall biosynthesis by inhibiting the hydroxy‐methylglutaryl coenzyme A (HMG‐CoA) reductase enzyme (HMGR), and lovastatin lactone can independently inhibit the activity of methylenetetrahydromethanopterin dehydrogenase (mtd), a key methanogenesis enzyme, by competing with coenzyme F420, a central catabolic cofactor required by mtd [14, 15]. Stool and small bowel (ileal) contents were collected at euthanasia and stored at − 80 °C prior to analysis.
Animal study design. A Investigation of the effects of diet-induced increases in the methanogen Methanobrevibacter smithii, and of methanogenesis inhibitors, on stool consistency. B Investigation of the effects of gavage with H2S producers Desulfovibrio piger and Fusobacterium varium on stool consistency
DNA Extraction
DNAs were extracted from stool and small bowel contents using the MagAttract PowerSoil DNA KF Kit (Qiagen) with some modifications [16]. Extracted DNAs were purified using a KingFisher Duo automated system (ThermoFisher Scientific, Waltham, MA), and DNA purity and concentration were determined using a NanoDrop One spectrophotometer (ThermoFisher Scientific).
Analysis of M. smithii Levels by Quantitative Polymerase Chain Reaction (qPCR)
Levels of the methanogen M. smithii in stool and small bowel contents were determined by qPCR using primers and probes targeting the beta subunit of the RNA polymerase (rpoB) gene [17]. Assays were optimized by Applied Biosystems (Custom Taqman Gene Expression Assays). Real-time qPCR was performed on a QuantStudio 6 Flex System (ThermoFisher Scientific) as follows: 1 µL of 20 × Custom TaqMan Gene Expression assay solution (ThermoFisher Scientific), 10 µL of TaqMan Fast Advanced Master Mix (ThermoFisher Scientific), 7 µL of PCR grade water and 2 µL of template DNA (25 ng/µl), at 50 °C for 2 min, 95 °C for 2 min, 40 cycles of 95 °C for 1 s, and 60 °C for 20 s. DNA from an M. smithii stock culture was extracted using the same protocol, and standard curves with tenfold serial dilutions were prepared for use as qPCR standards.
Effects of H2S Producers on Stool Consistency
The effects of H2S producers on stool consistency were determined using D. piger and F. varium. D. piger ATCC29098 (ATCC, Virginia) was grown anaerobically in sterile 1249 modified Baar’s medium for sulfate reducers (ATCC) and plated on trypticase soy agar (TSA) with 5% sheep blood (BD, New Jersey). Plates were incubated anaerobically at 37◦C for 48–96 h to obtain single colonies. H2S production was confirmed by growing isolated D. piger colonies in 1249 modified Baar’s medium for sulfate reducers with 5% of ferrous ammonium sulfate.
F. varium clinical isolates were obtained from the Cedars-Sinai Microbiology Department and grown anaerobically in sterile peptone yeast extract broth (Anaerobe Systems, Morgan Hill, CA) and plated on TSA with 5% sheep blood (BD). Isolated single F. varium colonies were cultured in SIM (Sulfide, Indole, Motility) medium (Hardy Diagnostics, CA) to confirm H2S production.
Gavage with H2S Producers
Rats were divided into three groups and gavaged with (1) sterile 1 × PBS (controls), (2) 1 × 108 CFU/mL D. piger, or (3) 1 × 108 CFU/mL F. varium. Liquid cultures were grown from single colonies as described above until 1 × 108 CFU/mL was achieved. Cultures were centrifuged for 10 min at 3000×g at 4 °C, and bacterial pellets were washed twice and resuspended in 1 × PBS to achieve 1 × 108 CFU/mL. Rats were gavaged on days 0, 2, and 4 (Fig. 1). These rats were fed a standard chow diet (13% energy from fat [PicoLab Rodent Diet 20; LabDiet, St. Louis, MO]).
Stool Collection and H2S Measurements
Stool samples were collected by anal stimulation [18] at baseline (3-day collection) and on days 5, 7, 10, 12, and 20 (single collections, see Fig. 1) for determining stool consistency (% wet weights). H2S production in stool from all rats was measured on day 20. Samples were immediately homogenized with sterile 1 × PBS, placed in sterile Erlenmeyer flasks sealed with rubber stoppers connected to a stopcock and incubated at 37 °C for 2 h. Gas samples were withdrawn into gas-impermeable bags and sent for measurement of gases by gas chromatography (Gemelli Biotech, Raleigh, NC).
16S rRNA Sequencing and Analysis
Stool samples collected at euthanasia from animals gavaged with H2S producers were used for DNA isolation and preparation of 16S rDNA hypervariable V3 and V4 region libraries following an Illumina protocol (https://support.illumina.com/documents/documentation/chemistry_documentation/16s/16smetagenomic-library-prep-guide-15044223-b.pdf) (Illumina, San Diego, CA), using primers S-D-Bact0341-b-S-17 and SD-Bact-0785-a-A-21 [19] as described previously [16]. Final libraries were analyzed using Agilent High Sensitivity DNA chips (Agilent) on an Agilent 2100 Bioanalyzer System and quantified using Qubit 1X dsDNA High Sensitivity Assay kits (Invitrogen by Thermo Fisher Scientific, Waltham, MA, USA) on a Qubit 4 Fluorometer (Invitrogen). Pooled libraries were paired-sequenced (2 × 301) on a MiSeq instrument (Illumina). Reads were trimmed and merged using CLC Genomics Workbench software version 22.0.2 (Qiagen). Operational Taxonomic Unit (OTU) clustering and taxonomic analyses were carried out using CLC Microbial Genomics Module 22.1.2 (Qiagen) against the SILVA Small Subunit (SSU) rRNA Database v. 138.1 (2020), with 97% of similarity and using default parameters. Creation of new OTUs was not allowed. Alpha diversity (Shannon index) and beta diversity (Unweighted UniFrac) were calculated using the CLC Microbial Genomics Module (Qiagen). The PERMANOVA test was used to calculate differences in beta diversity. Differences in microbial relative abundances between groups were calculated using the CLC Microbial Genomics Module (Qiagen). Significance was determined by Wald test and Wilcoxon test and, due to the small number of samples, a False Discovery Rate (FDR) < 0.1 was considered significant. Microbial metabolic pathways were analyzed using MetaCyc Pathway Database (2022–05) and CLC Microbial Genomics Module (Qiagen).
Statistical Analysis
Descriptive analyses are presented as mean ± standard deviation. Continuous variables were compared by t-test or Mann–Whitney U-test for comparisons between two groups. Comparisons between three or more groups were analyzed by one-way ANOVA or Kruskal–Wallis. Correlations between variables were calculated by Spearman’s test (two-tailed). ROC curves were used to analyze thresholds for H2S levels. Statistical analysis was performed using SPSS 24.0 (SPSS® Inc., Chicago, IL), and GraphPad Prism® 9.5.1 (GraphPad Software, La Jolla, CA). Graph construction was performed using GraphPad Prism® 9.5.1 (GraphPad Software). Significance was set at P < 0.05.
Results
Methanobrevibacter smithii Is Associated with a Constipation-Like Phenotype in Rats
To determine the effects of increased M. smithii levels on stool phenotypes, adult male Sprague Dawley rats (N = 30) were placed on a HFD. Absolute levels of stool M. smithii were increased significantly after 3 days on HFD when compared to baseline (1.69 × 105 ± 1.57 × 105 copies/gram vs. 0.91 × 105 ± 0.71 × 105 copies/gram, P = 0.0127) and were further increased after 52 days on HFD (3.06 × 105 ± 2.89 × 105 copies/gram, P < 0.001, Fig. 2A). Stool % wet weights were significantly decreased after 12 days on HFD compared to baseline (P < 0.0001) and remained significantly decreased after 59 days on HFD (P < 0.0001, Fig. 2B). Further, the decrease in % wet weight after 59 days on HFD was associated with the increase in M. smithii levels (R = − 0.38, P = 0.037), indicating that higher M. smithii levels were associated with drier stool.
Changes in absolute levels of M. smithii in stool (A) and stool % wet weights (B) of rats after 59 days on HFD. Changes in absolute levels of M. smithii in stool (C) and stool wet weights (D) after 10 days of treatment with water (controls), lovastatin hydroxyacid, or lovastatin lactone. E Changes in absolute levels of M. smithii in the small bowel (ileum) after 10 days of treatment with water (controls), lovastatin hydroxyacid, or lovastatin lactone. Horizontal bars denote mean ± SD
To confirm that these effects were due to increased M. smithii, rather than the effects of the HFD alone, rats were randomized into 3 groups and treated for 10 days with lovastatin lactone (N = 10), lovastatin hydroxyacid (N = 10), or water (controls, N = 10) while being maintained on the HFD. Levels of M. smithii in stool samples were not significantly different between controls and rats treated with either form of lovastatin (Fig. 2C). However, rats treated with lovastatin hydroxyacid exhibited a significant increase in % wet weight when compared to controls (P = 0.0246, Fig. 2D), indicating a partial reversal of the constipation-like phenotype.
In contrast to stool, rats treated with lovastatin lactone exhibited significant reductions in absolute M. smithii levels in the distal small bowel (ileum) (29.24 × 105 ± 19.91 × 105 copies/gram) when compared both to controls (144.80 × 105 ± 82.15 × 105 copies/gram, P < 0.0006) and to rats treated with lovastatin hydroxyacid (P = 0.0003, Fig. 2E). Levels of M. smithii in the small bowel of rats treated with lovastatin hydroxyacid were not significantly different from those in controls (Fig. 2E).
H2S-Producing Bacteria Induce H2S Production and a Diarrhea-Like Phenotype in Rats
To determine the effects of increased levels of H2S producers on stool phenotypes, adult male Sprague Dawley rats were gavaged with D. piger (N = 16), F. varium (N = 16) ,or PBS (controls, N = 8). Rats gavaged with D. piger had increased % wet weights on day 10 compared to controls (P < 0.0001, Fig. 3A). Rats gavaged with F. varium had significantly increased % wet weights at all time points when compared to controls [day 5 (P = 0.019), day 7 (P = 0.005), day 10 (P < 0.0001), day 12 (P = 0.027), and day 20 (P = 0.032)], with the peak occurring on day 10 (Fig. 3B). In addition, H2S production in stool samples from rats gavaged with D. piger (P = 0.0005) or F. varium (P = 0.006) was significantly greater than H2S production in stool samples from control rats (Fig. 3C). Using a cutoff of 0.48 ppm of H2S (95% Cl 0.65–1.0), stool H2S levels in rats gavaged with F. varium were positively associated with stool % wet weight on day 20 (r = 0.447, P = 0.042).
Changes in stool wet weight in rats gavaged with A D. piger and B F. varium. P-values denote comparisons between SRB-gavaged rats and controls. C H2S production in stool collected from D. piger- or F. varium-gavaged rats and controls. Horizontal bars denote mean ± SD. Statistical analyses by Mann–Whitney U-test
H2S Producers Induce Changes in Stool Microbial Profiles
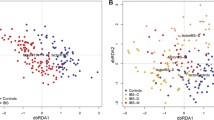
16S rRNA gene sequencing was performed using stool samples obtained at euthanasia, as rats gavaged with H2S producers exhibited increased stool H2S levels when compared to controls at this timepoint. Alpha diversity analysis (Shannon index) showed no differences between rats gavaged with H2S producers and controls, D. piger vs. controls, P = 0.5; F. varium vs. controls, P = 0.2). Although D. piger and F. varium were not detected in stool samples at euthanasia (17 days after the last gavage) and alpha diversity was not different between groups, there were significant differences in the stool microbial profiles of rats gavaged with H2S producers when compared to controls. Beta diversity (PERMANOVA) was different between control rats and rats gavaged with D. piger (P = 0.0001) and between control rats and rats gavaged with F. varium (P = 0.001) (Fig. 4). Moreover, beta diversity was positively associated with stool H2S levels (PCo3: r = − 0.401, P = 0.014).
Taxonomic differences were identified even at the phylum level between rats gavaged with H2S producers and controls, including increases in relative abundance (RA) of Deferribacterota (D. piger vs. controls, fold change (FC) = 8.93, P = 0.0006, Adj. P-value = 0.006; F. varium vs. controls, FC = 7.70, P = 0.002, Adj. P-value = 0.007), Bacteroidota (D. piger vs. controls, FC = 1.58, P = 0.002, Adj. P-value = 0.01; F. varium vs. controls, FC = 1.72, P = 0.0003, Adj. P-value = 0.004), Desulfobacterota (D. piger vs. controls, FC = 2.13, P = 0.017, Adj. P-value = 0.06; F. varium vs. controls, FC = 2.61, P = 0.003, Adj. P-value = 0.008). F. varium-gavaged rats also exhibited increased RA of phylum Firmicutes compared to controls (FC = 1.78, P = 0.002, Adj. P-value = 0.007).
At the family level, rats gavaged with D. piger exhibited increased RA of Bacteroidaceae (FC = 1.83, P < 0.0001, Adj. P-value = 0.002), Deferribacteraceae (FC = 10.15, P = 0.0005, Adj. P-value = 0.015), Lachnospiraceae (FC = 2.21, P = 0.0007, Adj. P-value = 0.015), Enterobacteriaceae (FC = 7.12, P = 0.001, Adj. P-value = 0.016), Staphylococcaceae (FC = 4.54, P = 0.003, Adj. P-value = 0.032), and Rikenellaceae (FC = 1.60, P = 0.01, Adj. P-value = 0.074), and decreased RA of Eggerthellaceae (FC = − 1.52, P = 0.006, Adj. P-value = 0.055), when compared to controls. Rats gavaged with F. varium exhibited increased RA of family Butyricicoccaceae (FC = 2.24, P = 0.0004, Adj. P-value = 0.023) when compared to controls. However, at the genus level, the RA of several genera were found to be different in F. varium-gavaged rats compared to controls, including increased RA of an unknown genus from family Lachnospiraceae (FC = 19.94, P = 0.0008, Adj. P-value = 0.039) and of Bilophila (FC = 4.77, P = 0.001, Adj. P-value = 0.048), and decreased RA of Akkermansia (FC = − 19.03, P = 0.004, Adj. P-value = 0.076). D. piger-gavaged rats also exhibited increased RA of the unknown genus from family Lachnospiraceae (FC = 18.96, P = 0.0009, Adj. P-value = 0.052), as well as Frisingicoccus (FC = 106.70, P = 0.0005, Adj. P-value = 0.052) and Mucispirillum (FC = 6.86, P-value = 0.002, Adj. P-value = 0.069), and a trend toward increased RA of Bilophila (FC = 3.66, P = 0.007, Adj. P-value = 0.116) and Sutterella (FC = 31.47, P = 0.006, Adj. P-value = FDR P = 0.116) (Supplemental Table 1).
A Spearman correlation was performed, including all animals, between stool H2S levels and the RA of known H2S-producing bacteria, as well as the most abundant bacteria found in the stool of these animals. There were positive associations between stool H2S levels and the RA of genera Bilophila (r = 0.282, P = 0.091), Escherichia-Shigella (r = 0.315, P = 0.058), the unknown genus from family Lachnospiraceae (r = 0.28, P = 0.093), and Sutterella (r = 0.331, P = 0.046). The positive association between H2S levels and the unknown genus from family Lachnospiraceae (r = 0.49, P = 0.024) was stronger in rats gavaged with F. varium, whereas the association with Sutterella was stronger in rats gavaged with D. piger (r = 0.592, P-value = 0.003).
Predicted microbial metabolic pathways were also analyzed and compared to those in controls, and several pathways were found to be different between groups. Importantly, the L-homocysteine biosynthesis pathway, which is used by specific bacteria to consume and produce H2S, was positively associated with stool % wet weight on day 20 (r = 0.30, P = 0.06). This pathway was increased in rats gavaged with F. varium compared to controls (P < 0.001) and was positively associated with H2S levels in these animals (r = 0.5, P = 0.021), but this was not observed when D. piger-gavaged animals were analyzed (r = 0.24, P = 0.264).
Discussion
In this study, we demonstrate that induced increases in levels of the methanogen M. smithii result in a constipation-like phenotype (decreased stool % wet weights) in a rat model, whereas increases in either of the H2S-producing species D. piger or F. varium result in a diarrhea-like phenotype (increased stool % wet weights) and increased stool H2S production. These findings build on our recent human studies linking increased levels of stool methanogens, including M. smithii, to increased breath methane CH4 in IBS-C subjects [6], and linking increased abundances of H2S-producing taxa, including the genera Fusobacterium and Desulfovibrio, to increased breath H2S in IBS-D subjects [6]. Taken together, these results suggest that methanogens such as M. smithii, and H2S producers such as D. piger and F. varium, may contribute to the constipation-predominant and diarrhea-predominant subtypes in IBS subjects, respectively.
M. smithii and other hydrogenotrophic methanogens use H2 produced by syntrophic bacterial species for the generation of CH4 [20]. CH4 has been directly linked to slowing of intestinal transit in an animal model and to an increased motility index in CH4-producing IBS subjects [21]. Several studies have linked breath CH4 to constipation [22, 23] and to IBS-C [3, 24,25,26], and M. smithii has been shown to be the predominant methanogen in CH4-postive IBS-C subjects [8]. We recently found that increased CH4 on breath test correlated with increased abundance of methanogens, including M. smithii, in stool samples from IBS-C subjects, and predicted microbial metabolic pathway analysis indicated enrichment of pathways associated with methanogenesis in these subjects [6]. These findings are also consistent with an independent study which recently found that increased CH4 on breath test was associated with slower oroanal transit times in IBS subjects, whereas increased H2 on breath test was associated with more rapid oroanal transit times [11]. In the present study, increased stool M. smithii levels correlated with a constipation-like phenotype (decreased stool % wet weights) in a rat model. These rats were also treated with different forms of lovastatin, which have anti-methanogenesis effects based on their ability to compete with the key cofactor F420 for its binding site on the mtd enzyme (lactone form), as well as their ability to inhibit archaeal cell wall biosynthesis by inhibiting the HMGR enzyme (hydroxyacid form) [14, 15]. Treatment with lovastatin lactone resulted in a significant reduction in M. smithii levels in the distal small bowel (ileum), and treatment with lovastatin hydroxyacid resulted in looser stools, when compared to rats maintained on the HFD, indicating that inhibiting methanogens resulted in normalization of % wet weights away from the constipation-like phenotype. Of note, while methanogens are typically most abundant in the colon in humans, we have previously demonstrated high abundance of methanogens in the small intestine, both in rats [12] and in some human subjects [27]. These findings suggest that increased levels of methanogens, particularly M. smithii, directly contribute to a constipation phenotype in subjects with IBS-C.
H2S-producing bacteria use different substrates in the production of H2S—F. varium primarily uses the amino acid cysteine, whereas D. piger uses sulfate—and compete with methanogens for H2 [28]. In contrast to CH4, H2S is also produced by mammalian cells, albeit in lower amounts [29]. Although H2S is often considered to be a toxic gas (at least when present in excess [30]), in recent years it has been shown to be a signaling molecule involved in many physiological functions, with anti-inflammatory and anti-oxidative effects in lung injury [31] as well as beneficial effects in the cardiovascular system [32]. With regard to the gastrointestinal tract, we previously linked higher breath H2S levels to diarrhea [33], which is consistent with animal studies demonstrating that H2S acts as a smooth muscle relaxant, through the inhibition of L-type calcium channels [34]. In addition, H2S has been linked to the diarrheal disorder ulcerative colitis [35], and F. varium specifically has been identified in Japanese patients with ulcerative colitis [36, 37]. We recently identified higher relative abundances of genus Fusobacterium and Desulfovibrio species in IBS-D subjects, with strong correlations between Fusobacterium and H2S on breath test [6]. In the present study, we show for the first time that increased levels of F. varium or D. piger result in a diarrhea-like phenotype in a rat model, and an increase in stool production of H2S. These findings suggest that increased levels of H2S-producing bacteria may contribute to a diarrheal phenotype in subjects with IBS-D.
Interestingly, the 16S rRNA sequencing results did not identify the gavaged H2S producers in stool at the time of euthanasia. It is possible that colonization occurred in a more proximal part of the gut, or that colonization with the gavaged bacteria was more transient. However, gavage with H2S producers appears to result in increased RA of other H2S-producing bacteria in these animals. The RA of Bilophila, a well-known H2S producer [38], was increased in both rat models but particularly in rats gavaged with F. varium, whereas the RA of Sutterella, a sulfur metabolizing bacterium [39], was more significantly increased in rats gavaged with D. piger. Furthermore, Sutterella RA was also found to be associated with stool H2S levels. Rats gavaged with D. piger also exhibited an increase in family Lachnospiraceae. This bacterial family was recently shown to increase the production of reactive sulfur species, such as H2S, in the host [40]. We hypothesize that these bacteria could have taken a niche created by the H2S producers with which these rats were initially gavaged, although further work would be needed to prove this. Lastly, the L-homocysteine biosynthesis pathway, which is used to both consume and produce H2S [41], was positively associated with stool % wet weight on day 20 and was predicted to be increased in rats gavaged with F. varium compared to controls. Using a cutoff of 0.48 ppm H2S, we also identified a positive association between this pathway and stool H2S levels in rats gavaged with F. varium, which could explain the diarrhea-like phenotype in these animals.
This study has some limitations. M. smithii levels were induced indirectly through feeding a high-fat diet, rather than by a more direct method such as gavage. This approach was chosen as we have previously shown that gavage does not result in persistent increases in the abundance of M. smithii, which is strictly anaerobic, whereas significant and stable increases in M. smithii levels can be induced via a HFD [12]. The decreases in stool wet weights correlated with increases in M. smithii levels, and were partly reversible using methanogenesis inhibitors, suggesting that these effects were due to M. smithii rather than non-specific effects of the HFD. More studies are needed to study the mechanistic effects of these organisms and the gases that they produce in the gastrointestinal tract.
In conclusion, this study further substantiates the presence of distinct microbiome-based subtypes (microtypes) that drive the predominant bowel phenotypes in IBS, consistent with the recent suggestion by Algera et al. that IBS subjects with more rapid vs. slow oroanal times have different underlying gut microbial compositions and associated breath gas profiles [11]. Specifically, we show that increased levels of methanogenic archaea such as M. smithii appear to drive a constipation phenotype, and increases in H2S-producing bacteria such as F. varium or D. piger appear to drive a diarrheal phenotype. These findings, and an increased understanding of the underlying mechanisms, may facilitate the development of targeted microtype-based therapies in IBS.
References
Black CJ, Ford AC. Global burden of irritable bowel syndrome: trends, predictions and risk factors. Nat Rev Gastroenterol Hepatol. 2020;17:473–486.
Lacy BE, Mearin F, Chang L, Chey WD, Lembo AJ, Simren M et al. Bowel Disorders. Gastroenterology. 2016;150:1393–407.e5.
Pimentel M, Lembo A. Microbiome and Its Role in Irritable Bowel Syndrome. Dig Dis Sci. 2020;65:829–39.
Wang L, Alammar N, Singh R, Nanavati J, Song Y, Chaudhary R et al. Gut Microbial Dysbiosis in the Irritable Bowel Syndrome: A Systematic Review and Meta-Analysis of Case-Control Studies. J Acad Nutr Diet. 2020;120:565–586.
Liu HN, Wu H, Chen YZ, Chen YJ, Shen XZ, Liu TT. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: A systematic review and meta-analysis. Dig Liver Dis. 2017;49:331–337.
Villanueva-Millan MJ, Leite G, Wang J, et al. Methanogens and Hydrogen Sulfide Producing Bacteria Guide Distinct Gut Microbe Profiles and Irritable Bowel Syndrome Subtypes. Am J Gastroenterol. 2022.
Pimentel M, Lezcano S. Irritable Bowel Syndrome: Bacterial Overgrowth–What’s Known and What to Do. Curr Treat Options Gastroenterol. 2007;10:328–337.
Kim G, Deepinder F, Morales W, Hwang L, Weitsman S, Chang C et al. Methanobrevibacter smithii is the predominant methanogen in patients with constipation-predominant IBS and methane on breath. Dig Dis Sci. 2012;57:3213–3218.
Ghoshal U, Shukla R, Srivastava D, Ghoshal UC. Irritable Bowel Syndrome, Particularly the Constipation-Predominant Form, Involves an Increase in Methanobrevibacter smithii, Which Is Associated with Higher Methane Production. Gut Liver. 2016;10:932–938.
Pimentel M, Saad RJ, Long MD, Rao SSC. ACG Clinical Guideline: Small Intestinal Bacterial Overgrowth. Am J Gastroenterol. 2020;115:165–178.
Algera JP, Colomier E, Melchior C, Hreinsson JP, Midenfjord I, Clevers E et al. Associations between postprandial symptoms, hydrogen and methane production, and transit time in irritable bowel syndrome. Neurogastroenterol Motil. 2022;11:e14482.
Mathur R, Kim G, Morales W, Sung J, Rooks E, Pokkunuri V et al. Intestinal Methanobrevibacter smithii but not total bacteria is related to diet-induced weight gain in rats. Obesity (Silver Spring). 2013;21:748–754.
Pimentel M, Morales W, Jee SR, Low K, Hwang L, Pokkunuri V et al. Antibiotic prophylaxis prevents the development of a post-infectious phenotype in a new rat model of post-infectious IBS. Dig Dis Sci. 2011;56:1962–1966.
Gottlieb K, Wacher V, Sliman J, Pimentel M. Review article: inhibition of methanogenic archaea by statins as a targeted management strategy for constipation and related disorders. Aliment Pharmacol Ther. 2016;43:197–212.
Muskal SM, Sliman J, Kokai-Kun J, Pimentel M, Wacher V, Gottlieb K. Lovastatin lactone may improve irritable bowel syndrome with constipation (IBS-C) by inhibiting enzymes in the archaeal methanogenesis pathway. F1000Res. 2016;5:606.
Leite G, Morales W, Weitsman S, et al. Optimizing Microbiome Sequencing for Small Intestinal Aspirates: Validation of Novel Techniques through the REIMAGINE Study. BMC Microbiol. 2019;19:https://doi.org/10.1186/s12866-019-1617-1.
Dridi B, Henry M, El Khechine A, Raoult D, Drancourt M. High prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae detected in the human gut using an improved DNA detection protocol. PLoS One. 2009;4:e7063.
Pimentel M, Chatterjee S, Chang C, Low K, Song Y, Liu C et al. A new rat model links two contemporary theories in irritable bowel syndrome. Dig Dis Sci. 2008;53:982–989.
Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1.
Gaci N, Borrel G, Tottey W, O’Toole PW, Brugere JF. Archaea and the human gut: new beginning of an old story. World J Gastroenterol. 2014;20:16062–16078.
Pimentel M, Lin HC, Enayati P, van den Burg B, Lee HR, Chen JH et al. Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1089–G1095.
Pimentel M, Mayer AG, Park S, Chow EJ, Hasan A, Kong Y. Methane production during lactulose breath test is associated with gastrointestinal disease presentation. Dig Dis Sci. 2003;48:86–92.
Kunkel D, Basseri RJ, Makhani MD, Chong K, Chang C, Pimentel M. Methane on breath testing is associated with constipation: a systematic review and meta-analysis. Dig Dis Sci. 2011;56:1612–1618.
Chatterjee S, Park S, Low K, Kong Y, Pimentel M. The degree of breath methane production in IBS correlates with the severity of constipation. Am J Gastroenterol. 2007;102:837–841.
Hwang L, Low K, Khoshini R, Melmed G, Sahakian A, Makhani M et al. Evaluating breath methane as a diagnostic test for constipation-predominant IBS. Dig Dis Sci. 2010;55:398–403.
Triantafyllou K, Chang C, Pimentel M. Methanogens, methane and gastrointestinal motility. Journal of neurogastroenterology and motility. 2014;20:31–40.
Kim G, Giamarellos EJ, Pyleris E, et al. Methanobrevibacter smithii is found in human duodenum and is associated with altered luminal cytokines. Gastroenterology. 2012;142:S-98.
Smith NW, Shorten PR, Altermann EH, Roy NC, McNabb WC. Hydrogen cross-feeders of the human gastrointestinal tract. Gut Microbes. 2019;10:270–288.
Carbonero F, Benefiel AC, Alizadeh-Ghamsari AH, Gaskins HR. Microbial pathways in colonic sulfur metabolism and links with health and disease. Front Physiol. 2012;3:448.
Doujaiji B, Al-Tawfiq JA. Hydrogen sulfide exposure in an adult male. Ann Saudi Med. 2010;30:76–80.
Zimmermann KK, Spassov SG, Strosing KM, Ihle PM, Engelstaedter H, Hoetzel A et al. Hydrogen Sulfide Exerts Anti-oxidative and Anti-inflammatory Effects in Acute Lung Injury. Inflammation. 2018;41:249–259.
Skovgaard N, Gouliaev A, Aalling M, Simonsen U. The role of endogenous H2S in cardiovascular physiology. Curr Pharm Biotechnol. 2011;12:1385–1393.
Singer-Englar T, Rezaie A, Gupta K, et al. 182 - Competitive Hydrogen Gas Utilization by Methane- and Hydrogen Sulfide-Producing Microorganisms and Associated Symptoms: Results of a Novel 4-Gas Breath Test Machine. Gastroenterology. 2018;154:S-47.
Quan X, Luo H, Liu Y, Xia H, Chen W, Tang Q. Hydrogen sulfide regulates the colonic motility by inhibiting both L-type calcium channels and BKCa channels in smooth muscle cells of rat colon. PLoS One. 2015;10:e0121331.
Linden DR. Hydrogen sulfide signaling in the gastrointestinal tract. Antioxid Redox Signal. 2014;20:818–830.
Ohkusa T, Okayasu I, Ogihara T, Morita K, Ogawa M, Sato N. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. 2003;52:79–83.
Yukawa T, Ohkusa T, Shibuya T, Tsukinaga S, Mitobe J, Takakura K et al. Nested Culture Method Improves Detection of Fusobacterium from Stool in Patients with Ulcerative Colitis. Japanese Journal of Infectious Diseases. 2013;66:109–114.
Peck SC, Denger K, Burrichter A, Irwin SM, Balskus EP, Schleheck D. A glycyl radical enzyme enables hydrogen sulfide production by the human intestinal bacterium Bilophila wadsworthia. Proc Natl Acad Sci U S A. 2019;116:3171–3176.
Nguyen LH, Cao Y, Hur J, et al. The Sulfur Microbial Diet Is Associated With Increased Risk of Early-Onset Colorectal Cancer Precursors. Gastroenterology. 2021;161:1423–32 e4.
Uchiyama J, Akiyama M, Hase K, Kumagai Y, Kim YG. Gut microbiota reinforce host antioxidant capacity via the generation of reactive sulfur species. Cell Rep. 2022;38:110479.
Soda K. Microbial sulfur amino acids: an overview. Methods Enzymol. 1987;143:453–459.
Acknowledgments
We would like to thank Dr. Margie Morgan (Department of Microbiology, Cedars-Sinai Medical Center) for providing us with the Fusobacterium varium strain, and Dr. Robert Gunsalus (Department of Microbiology, Immunology & Molecular Genetics, University of California, Los Angeles) for providing the Methanobrevibacter smithii. We would also like to thank the following for their support of the MAST research program: the John and Geraldine Cusenza Family Foundation, the Tull Family Foundation, the Monica Lester Charitable Trust, the Elias, Genevieve, and Georgianna Atol Charitable Trust, the National Philanthropic Trust, Synthetic Biologics, Inc, and Bausch Health. We would also like to thank Frank Lee and Joel Levine for their ongoing research support.
Funding
Open access funding provided by SCELC, Statewide California Electronic Library Consortium. This study was supported in part by funds from The Monica Lester Charitable Trust (RM), The Elias, Genevieve, and Georgianna Atol Charitable Trust (RM), and The National Philanthropic Trust (MP).
Author information
Authors and Affiliations
Contributions
Conceptualization: MP, AR, Formal analysis: GL, MJVM, AR, MP, Methodology: MJVM, GL, WM, SW, MP, Investigation: MJVM, GL, GP, GMB, MS, SC, DC, SW, HD, AR, MP, Visualization: MJVM, GL, Funding acquisition: GB, RM, MP, Project administration: RM, MP, Supervision: WM, SW, RM, MP, Writing—original draft: MJVM, GL, GB, WM, MP, Writing—review and editing: MJVM, GL, GB, WM, AR, RM, MP.
Corresponding author
Ethics declarations
Conflict of interest
M.P. is a consultant for Bausch Health, Ferring Pharmaceuticals Inc., and Vivante Health Inc. M.P. has received grant support from Bausch Health and Synthetic Biologics. R.M. has received grant support from Valiant Pharmaceuticals. A.R. is a consultant/speaker for and has received grant support from Bausch Health. Cedars-Sinai has a licensing agreement with Gemelli Biotech. A.R., M.P., and R.M. have equity in Gemelli Biotech. All other authors report no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
An invited commentary on this article is available at https://doi.org/10.1007/s10620-023-08198-4.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, which permits any non-commercial use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc/4.0/.
About this article
Cite this article
Villanueva-Millan, M.J., Leite, G., Morales, W. et al. Hydrogen Sulfide Producers Drive a Diarrhea-Like Phenotype and a Methane Producer Drives a Constipation-Like Phenotype in Animal Models. Dig Dis Sci 69, 426–436 (2024). https://doi.org/10.1007/s10620-023-08197-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-023-08197-5