Abstract
Background and Aims
Current genetic research of nonalcoholic steatohepatitis (NASH) cirrhosis is limited by our ability to accurately identify cases on a large scale. Our objective was to develop and validate an electronic health record (EHR) algorithm to accurately identify cases of NASH cirrhosis in the EHR.
Methods
We used Clinical Query 2, a search tool at Beth Israel Deaconess Medical Center, to create a pool of potential NASH cirrhosis cases (n = 5415). We created a training set of 300 randomly selected patients for chart review to confirm cases of NASH cirrhosis. Test characteristics of different algorithms, consisting of diagnosis codes, laboratory values, anthropomorphic measurements, and medication records, were calculated. The algorithms with the highest positive predictive value (PPV) and the highest F score with a PPV ≥ 80% were selected for internal validation using a separate random set of 100 patients from the potential NASH cirrhosis pool. These were then externally validated in another random set of 100 individuals using the research patient data registry tool at Massachusetts General Hospital.
Results
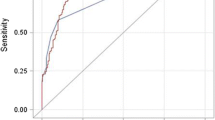
The algorithm with the highest PPV of 100% on internal validation and 92% on external validation consisted of ≥ 3 counts of “cirrhosis, no mention of alcohol” (571.5, K74.6) and ≥ 3 counts of “nonalcoholic fatty liver” (571.8–571.9, K75.81, K76.0) codes in the absence of any diagnosis codes for other common causes of chronic liver disease.
Conclusions
We developed and validated an EHR algorithm using diagnosis codes that accurately identifies patients with NASH cirrhosis.

Similar content being viewed by others
References
Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res.. 2014;42(D1):D1001–D1006.
Visscher PM, Wray NR, Zhang Q, et al. 10 Years of GWAS discovery: biology, function, and translation. Am J Hum Genet.. 2017;101(1):5–22.
Dawber TR, Meadors GF, Moore FE. Epidemiological approaches to heart disease: the Framingham study. Am J Public Health.. 1951;41(3):279–281.
Wei WQ, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med.. 2015;7(1):1–14.
Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Informa Assoc.. 2012;19(2):212–218.
Blumenthal D, Tavenner M. The, “meaningful use” regulation for electronic health records. N Engl J Med.. 2010;363(6):501–504.
Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc.. 2013;20(E1):147–154.
Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet.. 2011;89(4):529–542.
Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet.. 2010;86(4):560–572.
Kurreeman F, Liao K, Chibnik L, et al. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet.. 2011;88(1):57–69.
Beste LA, Leipertz SL, Green PK, et al. Trends in burden of cirrhosis and hepatocellular carcinoma by underlying liver disease in US Veterans, 2001–2013. Gastroenterology.. 2015;149(6):1471–1482.e5.
Corey KE, Kartoun U, Zheng H, Shaw SY. Development and validation of an algorithm to identify nonalcoholic fatty liver disease in the electronic medical record. Dig Dis Sci.. 2016;61(3):913–919.
Matteoni CZY, Gramlich T, et al. Nonalcoholic fatty liver disease: a spectrum of clinical and pathological severity. Gastroenterology. 1999;116(6):1413–1419.
Galle PR, Forner A, Llovet JM, et al. EASL clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol.. 2018;69(1):182–236.
Younossi ZM, Gramlich T, Liu YC, et al. Nonalcoholic fatty liver disease: assessment of variability in pathologic interpretations. Mod Pathol.. 1998;11(6):560–565.
Younossi Z, Stepanova M, Sanyal AJ, et al. The conundrum of cryptogenic cirrhosis: adverse outcomes without treatment options. J Hepatol.. 2018;69(6):1365–1370.
Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc.. 2013;20(e1):e147–e154.
Liao KP, Cai T, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken).. 2010;62(8):1120–1127.
van der Ploeg T, Austin PC, Steyerberg EW. Modern modelling techniques are data hungry: a simulation study for predicting dichotomous endpoints. BMC Med Res Methodol.. 2014;14:137.
Husain N, Blais P, Kramer J, et al. Nonalcoholic fatty liver disease (NAFLD) in the Veterans Administration population: development and validation of an algorithm for NAFLD using automated data. Aliment Pharmacol Ther.. 2014;40(8):949–954.
Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med.. 2015;12(3):1–10.
Borodulin K, Tolonen H, Jousilahti P, et al. Cohort profile: the national FINRISK study. Int J Epidemiol.. 2018;47(3):696–1471.
Rich NE, Oji S, Mufti AR, et al. Racial and ethnic disparities in nonalcoholic fatty liver disease prevalence, severity, and outcomes in the united states: a systematic review and meta-analysis. Clin Gastroenterol Hepatol.. 2018;16(2):198–210.e2.
Teixeira PL, Wei W-Q, Cronin RM, et al. Evaluating electronic health record data sources and algorithmic approaches to identify hypertensive individuals. J Am Med Inform Assoc.. 2017;24(1):162–171.
Delaney JT, Ramirez AH, Bowton E, et al. Predicting clopidogrel response using DNA samples linked to an electronic health record. Clin Pharmacol Ther.. 2012;91(2):257–263.
Bowton E, Field JR, Wang S, et al. Biobanks and electronic medical records: enabling cost-effective research. Sci Transl Med. 2014;6(234):234cm3.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No outside funding or grant support was involved in the production of this manuscript. The authors have no pertinent conflicts of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Danford, C.J., Lee, J.Y., Strohbehn, I.A. et al. Development of an Algorithm to Identify Cases of Nonalcoholic Steatohepatitis Cirrhosis in the Electronic Health Record. Dig Dis Sci 66, 1452–1460 (2021). https://doi.org/10.1007/s10620-020-06388-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-020-06388-y