Abstract
Small island populations are vulnerable to genetic decline via demographic and environmental stochasticity. In the absence of immigration, founder effects, inbreeding and genetic drift are likely to contribute to local extinction risk. Management actions may also have a greater impact on small, closed populations. The demographic and social characteristics of a species can, however, delay the impact of threats. K’gari, a ~ 1 660 km2 island off the Australian east coast and UNESCO World Heritage Site (Fraser Island 1842–2023), supports an isolated population of approximately 70–200 dingoes that represent an ideal opportunity to explore the small island paradigm. To examine temporal and spatial patterns of genetic diversity in this population we analysed single nucleotide polymorphism (SNP) genotype data (72 454 SNPS) for 112 K’gari dingoes collected over a 25-year period (1996 to 2020). Genetic diversity was lower in K’gari dingoes than mainland dingoes at the earliest time point in our study and declined significantly following a management cull in 2001. We did not find any spatial genetic patterns on the island, suggesting high levels of genetic connectivity between socially discrete packs. This connectivity, combined with the social structure and behaviour of dingoes, may act in concert to buffer the population from the impacts of genetic drift in the short term. Nevertheless, a general decline in genetic variation via inbreeding and drift has occurred over the past 20 years which we suggest should be considered in any future management planning for the population. Monitoring patterns of genetic variation, together with a clearer understanding of the social ecology of K’gari dingoes, will aid in the development of measurable genetic targets set over ecologically meaningful timelines, and help ensure continued survival of this culturally important population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Small populations of wildlife are generally more vulnerable to stochastic perturbations than larger ones (MacArthur and Wilson 1963; Caughley 1994). Endemic and island populations typify the increased extinction risk of the small and declining population paradigms (Pimm 1991; Frankham 1998), while extinction risks are generally higher on smaller islands (MacArthur and Wilson 1963). Inbreeding depression may also be an important contributing risk factor, particularly in naturally outbreeding species where there is compelling evidence that loss of genetic diversity through inbreeding contributes significantly to local extinction risk through reduced population fitness (Brook et al. 2002; Frankham 2005). But such effects are also likely to impact some species more than others (Elgar and Clode 2001). Increased inbreeding is also predicted in island populations when there are bottleneck effects at founding, together with subsequent low effective population sizes (Frankham 1996; Frankham and Ralls 1998). In the ongoing absence of migration, island populations are thus particularly susceptible to the negative effects of enhanced genetic drift (Vellend and Geber 2005).
While threshold effects associated with inbreeding depression may vary between populations and species (Frankham 1995; Hedrick 1999), Bouzat (2010) has argued that this ‘detrimental paradigm’ of population bottlenecks, isolation, and inbreeding depression is far more nuanced, depending largely on elements of selection, chance, human impact, and population history to explain the range of observed outcomes (e.g. Arauco-Shapiro et al. 2020; Gooley et al. 2017; Meiring et al. 2022; Ørsted et al. 2019; Torres-Florez et al. 2014). Moreover, genetic variation and fitness declines in small, isolated populations can also be tempered by their demographic characteristics (e.g., long generation time and overlapping generations, (Lippe et al. 2006; Bishop et al. 2009) and the social structure of species (Parreira and Chikhi 2015; Leach et al. 2020). Remnant populations may therefore retain genetic diversity for longer than expected for many reasons; but while the predicted effects of local inbreeding may be delayed, there is likely to be little forewarning of critical inbreeding risk thresholds in isolated populations (Frankham 1995).
Dingoes/wongari (hereafter dingoes) Canis familiaris (following Jackson et al. 2021, but also see Smith et al. 2019) are highly adaptable generalists, functioning as top-order predators across their range on the Australian mainland and a few offshore islands (Letnic et al. 2012). Their social structure is characterised by high flexibility and complex dynamics akin to other social canids, and centers on familial packs, each with a dominant male and female who account for most of the successful breeding (Corbett 1988). Across Australia dingoes have a long and divisive history, with conflicting opinions on (not exhaustively) what constitutes a dingo, their conservation value, their ecological importance, and how they should be managed (see Archer-Lean et al. 2015; Donfrancesco et al. 2023 for further discussion). Irrespective, dingoes are the largest terrestrial predator across mainland Australia and have been both an ecological and human-commensal element of the landscape with significant cultural importance to Traditional Custodians (Corbett 2001; Archer-Lean et al. 2015) for at least 3500 years based on archeological evidence, with genetic evidence suggesting 7000–11 000 years (Smith 2015; Cairns and Wilton 2016; Balme et al. 2018; Zhang et al. 2020; Bergström et al. 2020; Cairns 2021). Recent molecular evidence supports dingoes as a unique differentiated lineage from both domestic dogs and wolves (Cairns et al. 2022; Ballard et al. 2023); while nuclear, mitochondrial, and genomic data reveal signatures of at least two, and potentially four, broadly distributed dingo clades across Australia (Cairns et al. 2017, 2018, 2023; Stephens et al. 2022). Due to occasional hybridization between dingoes and domestic dogs first introduced in the late 18th century under British colonial rule, some contemporary dingo populations reveal a degree of domestic dog ancestry. Even so, most populations maintain a dingo-dominant genetic signature and large areas support ‘pure’ dingoes, particularly away from dense human settlements (Stephens et al. 2015, 2022; Cairns et al. 2022, 2023).
K’gari (previously known as Fraser Island between 1842 and 2023) sits off Australia’s east coast, where it forms part of the Great Sandy National Park and is the world’s largest sand island (~ 1 660 km2) with some sections only ~ 1 km offshore. The island has a long history of Aboriginal presence and was permanently disconnected from the mainland ~ 6 000 years bp due to rising sea levels in the mid-Holocene (Gontz et al. 2015). Whilst K’gari’s island status coincides with the outer boundaries of when dingoes may have arrived in the region, 3500–11000 years bp; it is possible that some of the original founder population may have been in-situ as the island became separated from the mainland. However, the island’s Traditional Custodians, the Butchulla People, are likely the main source of the founding population of dingoes on K’gari (Conroy et al. 2021). The island was also settled in post-colonial times for cattle and sheep farming, together with extensive timber extraction that finally ceased in the late 20th century (Petrie 1995; Walker et al. 2022). K’gari was designated a UNESCO World Heritage Site in 1992 and is a popular tourist destination collaboratively managed by the Queensland Parks and Wildlife Service (QPWS) and the Butchulla Aboriginal Corporation (BAC), who represent the longstanding cultural and spiritual connections of the Butchulla people with the island and its dingoes (Carter et al. 2017).
In this study we explore the effects of historic isolation together with recent population management (2001–2020) on genetic variation in the small population of dingoes on K’gari. K’gari dingoes are low in genetic diversity and are differentiated from mainland dingoes (Cairns et al. 2018; Conroy et al. 2021); the island is also an important reservoir of ‘pure’ dingoes because of minimal introgression from modern domestic dog breeds (Cairns et al. 2022). Using a single nucleotide polymorphism (SNP) dataset representing 25 years of K’gari dingoes and associated individual information we determine the longer-term effects of historic isolation and management actions on (i) the spatial distribution of genetic clusters on K’gari, (ii) temporal changes in genetic clusters, and (iii) temporal patterns of allelic richness, inbreeding and relatedness across the sampling period. K’gari dingoes provide an excellent case study to explore the genetic consequences of both historical geographic isolation and contemporary management strategies in a social species of substantial cultural and ecological significance.
Methods
Ethics statement
All relevant procedures were approved under existing legislation and/or authorised by the Queensland Department of Environment and Resource Management (DERM), Department of Environment and Science (DES), and Department of Agriculture and Fisheries (DAF) Animal Ethics Committee (permit numbers: DERM/2009/04/03, DERM2011/09/04, DERM2011/12/06, SA2012/12/404, SA 2015/11/532, SA 2018/12/672), and the study was conducted in accordance with these approvals.
Study site and management interventions
K’gari is situated within the Great Sandy National Park located off the south-east coast of Queensland, Australia (24°42′S, 153°15′E; Fig. 1). At 122 km long it is the world’s largest sand island, supporting half the world’s perched freshwater dune lakes, and characterised by extensive subtropical vegetation and beaches on its east coast. The current population of dingoes on K’gari is estimated to be 70–197 individuals (Allen et al. 2015). They have shared mitochondrial ancestry with dingoes from southeast Australia (Cairns et al. 2016), however Y-chromosome and genome-wide SNP analysis suggests they are a genetically distinct lineage and more closely related to dingoes from northwest Australia (Cairns et al. 2017; 2018).
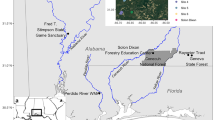
Map of K’gari and adjacent mainland showing the location of dingo samples used in this study. The four QPWS management regions, or research ‘base-areas’, used in the analysis below are delineated. Photos are of K’gari dingoes. Photo credit: top and bottom left are Linda Behrendorff and bottom right is Jacqueline Bishop
Currently, the island is divided into four spatial management units: Waddy Point, Dundubarra, Central and Eurong (Fig. 1). Historically, dingoes were occasionally removed from the island by managers (approximately 10 individuals between 1990 and 2000), but a policy of lethal management of K’gari dingoes was implemented in 2001 following the death of a nine-year-old child from an attack by dingoes. Between 2001 and 2013, 110 individuals were removed; approximately two thirds of these were young males (Allen et al. 2015), and occasional removals still occur in response to dingo aggression towards people (Behrendorff 2021). Ongoing management actions also include exclusion of dingoes from some human visitation sites such as some townships and campgrounds, and all rubbish disposal facilities using fencing and gates (Allen et al. 2015; Behrendorff 2021).
Sample source, selection, and genotyping
Samples used in this study were collected during routine management activities by Queensland Parks and Wildlife Service (QPWS) and represent the period 1996 to 2020. Ear tissue samples were collected opportunistically across the island from dead and killed dingoes. Individuals were selected from the QPWS sample archive (n = 300 samples available) for genetic analysis to ensure broad temporal and spatial coverage of the island based on the available metadata (n = 137 samples chosen for genotyping based on location at death and management area frequented by the individual; Fig. 1; Table 1, Online Resource 1). An additional seven tissue samples were obtained from dingoes at Rainbow Beach (~ 10 km south of K’gari) in 2019–2020 (Fig. 1) as representatives of adjacent mainland individuals.
DNA extraction and genotyping were performed by the Australian Genome Research Facility (AGRF, Adelaide, Australia) as follows: Genomic DNA was extracted using the DNeasy® Blood & Tissue kit (Qiagen, Germany) following manufacturer’s instructions. Quality assessment was performed by QantiFluor and samples normalised to approximately 200 ng in 4 µl. Samples were processed on the Illumina 170 K CanineHD Whole-Genome Genotyping BeadChip (Illumina Inc., San Diego, USA) following the Illumina Infinium HD Ultra Assay (AGRF NATA scope GGTMN00263). This SNP array includes 173 662 single nucleotide polymorphisms (SNPs) from across the domestic dog genome. Genotypes were called with Illumina’s GenomeStudio 2.0.4 with Genotyping module 2.0.4 software, using the default Illumina settings and Illumina CanineHD_B manifest and CanineHD_A cluster files were used for the analysis.
All analyses reported below were performed in R v4.1.2 in RStudio 2022.07.2 (R Core Team 2022; RStudio Team 2022) unless indicated otherwise. The tidyverse package was used extensively for data manipulation (Wickham et al. 2019). Unless indicated otherwise, ggplot2 (Wickham 2016) was used for visualisations. PLINK v1.9 was implemented in R and analyses were followed as explained on the PLINK 1.9 website (cog-genomics.org/plink/1.9) (Purcell et al. 2007; Purcell 2022). Relevant PLINK command flags have been included in each analysis.
Data processing - quality control
The dataset of 137 K’gari and seven Rainbow Beach dingoes was combined with data for 24 dingoes from Cairns et al. (2018) also analysed with the same SNP array. These 24 dingoes originated from across mainland Australia (representing both the southeast and northwest lineages) together with four individuals from K’gari (Cairns et al. 2018). All SNPs were aligned to the CanFam 3.1 genome (Lindblad-Toh et al. 2005) and quality control filtering was performed in PLINK v1.9. Individuals missing more than 20% of SNPs were excluded (25 K’gari samples) together with SNP loci with 20% missing data and a minor allele frequency (MAF) of less than 1%. SNPs with evidence of deviation from Hardy–Weinberg equilibrium that exceeded p < 1 × 10−6 were also excluded (Marees et al. 2018). This resulted in a dataset with 73 366 SNPs for 143 individuals (112 K’gari, 7 Rainbow Beach, 24 Cairns et al. (2018) with 4 from K’gari, Online Resource 2). SNPs on X and Y chromosomes and mitochondrial SNPs were then removed to avoid any bias caused by inheritance differences between males and females, resulting in 72 454 SNPs in the final dataset (PLINK commands and details in Online Resource 3). This dataset was used for inbreeding and homozygosity analyses, detailed below.
Data processing - linkage disequilibrium
Principal Coordinate Analysis (PCoA), clustering (PopCluster and TESS) and pairwise relatedness analyses are sensitive to linked markers (Abdellaoui et al. 2013) and thus the SNPs were pruned for these analyses to reduce linkage. The independent pairwise command in PLINK was implemented with a step size of 1500 kb and an r2 threshold of 0.2 (--indep-pairwise 1500 kb 1 0.2) resulting in 12 678 SNPs.
Temporal partitioning of SNP data
To explore temporal patterns in the distribution of genetic variation, we divided the 25-year dataset into five-year time periods (Periods A to E). Five years was selected because it represents 2–3 dingo generations (Lord et al. 2013) and provides a snapshot of the likely number of generations present in the population at any given time. The end of the first time period coincided with the management cull in 2001, allowing us to compare the population before and after this event. Given that all samples came from deceased animals, we had an accurate year of death. By combining this with known or estimated birth years, we were able to determine which dingoes were likely alive in each time period (Table 1; Online Resource 1).
Population structure - K’gari vs. mainland
The relationship between the K’gari and mainland populations was explored using two complimentary genetic clustering approaches, PCoA and PopCluster. Both analyses used the pruned dataset (12 678 SNPs). PCoA analyses was performed using the cmdscale command in base R (Meszaros 2021). The distance matrix was created in PLINK using the --distance-matrix option. Population structure analysis was performed using PopCluster v1.2.0.0 with the admixture model and unequal prior allele frequencies (Wang 2022). PopCluster (Wang 2022) uses likelihood methods for fast and accurate population admixture inference to identify population clustering. Twenty runs each for K values from 2 to 6 were performed. The best run, as identified by the program, was used for visualisations in R. The second order rate of change of the estimate log-likelihood (DLK2) and the mean of FST/FIS (FSTIS estimator) were used to assist in choosing the most representative value of K (Wang 2022).
K‘gari population - allelic diversity over time
Allelic diversity with the K’gari population was assessed over time by calculating the minor allele frequency (maf) within each time period using the --freq command in PLINK. ANOVA and Tukey tests were performed in R (aov; TukeyHSD in base R) to test for any significant differences between the five time periods.
K’gari population - inbreeding estimation over time
Inbreeding within the K’gari population over time was assessed using two complimentary methods, individual inbreeding coefficients and individual homozygosity. We calculated individual inbreeding coefficients (F; coancestry inbreeding) using the --het command in PLINK (Purcell et al. 2007). This F value is a relative comparison between observed and expected homozygosity between the sample and the whole population. Individual homozygosity was also reported (H; observed homozygosity per individual) from the --het output (Purcell et al. 2007). These analyses used the larger dataset (72 454 SNPs). Data were clustered by the dingoes born in the five time periods (Table 1) and compared to the mainland samples using box and whisker plots. ANOVA and Tukey tests were performed in R to test for any significant differences between time periods and the mainland.
K’gari population - spatial structure
Two complimentary analyses were used to look for any spatial structure within the K’gari population, TESS and PopCluster by base area. Both analyses used the pruned dataset (12 678 SNPs). TESS (tessr3 package in R) is a spatially informed population structure analysis (Caye and Francois 2016; Caye et al. 2016). This analysis was performed on the K’gari dataset with no separation into time periods to look for any genetic clustering across the island. K values from 2 to 12 were examined. Cross validation scores were plotted for each value of K to determine which number of ancestral populations was most likely. Visualisation of K values from 2 to 6 were prepared. PopCluster was run on the K’gari samples only as above (20 runs of each K from 2 to 10). Data were organised into the four management areas indicated in Fig. 1 to look for any broad spatial clustering.
K’gari population - genetic isolation over time
Population structure of the K’gari population over time was investigated using two complimentary approaches, PCoA and PopCluster. All analyses used the K’gari individuals only (n = 112) in the pruned dataset (12 678 SNPs). PCoA was performed as above. To explore differences between time periods (e.g. before and after management interventions), individuals were coded according to the time period in which they were born. A PERMANOVA was performed on the Eigenvalues from the PCoA (adonis2 in vegan package) followed by a Tukey test (pairwise.adonis2 in pairwiseAdonis package) to test for differences between time periods (Arbizu 2017; Oksanen et al. 2020).
PopCluster results from K’gari population - spatial structure above were rearranged to show the sampled animals alive in each of the five time-periods outlined in Table 1 and by birth year within each time period. Data were plotted for K = 2 to K = 4. Further analysis of the best run from K = 3 was performed: individuals within each time period were sorted by ‘Q’ values and plotted using a box and whisker plot. MANOVA indicated significant differences and thus ANOVA, followed by Tukey tests of each cluster, was performed to determine which time periods were significantly different from one another.
K’gari population - fine scale temporal analysis of relatedness estimates
Pairwise relatedness estimates (Wang’s estimator) were calculated for all K’gari individuals (n = 112) using the pruned dataset (12 678 SNPs) in Coancestry v1.0.1.9 (Wang 2011). A box and whisker plot of all pairwise relatedness estimates was generated; relationships between individuals alive in each time period (Table 1) were included in each cluster. A heat map of all pairwise relatedness estimates was plotted with individuals ordered based on birth year. ANOVA and Tukey tests were performed in R to determine whether relatedness levels varied significantly between time periods.
Results
Genotypes from 143 dingoes were received from AGRF (Online Resource 2). Filtering reduced the dataset to 119 individuals (112 from K’gari and 7 from Rainbow Beach) with 72 454 SNPs. Removal of linked SNPs further reduced the dataset to 12 678 SNPs (Online Resource 3).
Population structure - K’gari vs. mainland
The K’gari population was distinct from the mainland dingoes according to both the PCoA and PopCluster analyses (Fig. 2; Online Resource 4). The first two axes of the PCoA explained 47.36% of the variation in the data, with PC1 separating mainland dingoes from K’gari dingoes and PC2 separating the mainland dingo populations from each other (Fig. 2). K’gari individuals separated from the mainland samples at K = 2, although there was evidence of minimal mainland representation in the K’gari individuals in the 1996–2001 time period (Online Resource 4).
K‘gari population - allelic diversity over time
Allelic richness declined from an average of 6.60% in time period A down to 5.22% in time period E (A: 6.60%, B: 5.48%, C: 5.44%, D: 5.26%, E: 5.22%. ANOVA indicated the decline was significant (ANOVA: df = 4, F = 2.2 × 10−16). Time period A was significantly higher than all other time periods and there was a significant decline between time period B and time period D (Tukey test Table 2).
K’gari population - inbreeding estimation over time
Based on both analytical approaches used, K’gari individuals had significantly higher individual inbreeding (F) and homozygosity (H) values compared to mainland individuals. F values were significantly different (ANOVA: df = 5, F = 114.7, P < 2 × 10−16; Fig. 3a) as were H values (ANOVA: df = 5, F = 91.08, P < 2 × 10−16; Fig. 3c). Mean, standard error, minimum and maximum values per time period for both individual inbreeding (mean range: 0.30–0.46 on K’gari; mainland: − 0.55) and H (mean range: 64 107–66 781 on K’gari; mainland: 56 750) are reported in Online Resource 5.
Measures of inbreeding within the K’gari population and in comparison to mainland dingo individuals (based on 72 454 SNPs); “main” includes the mainland genotypes reported in Cairns et al. (2018) and the Rainbow Beach genotypes from this study as a comparison; A–E represent the time periods as per Table 1; a F values over time as calculated in PLINK; b Tukey test results for statistically significant differences between the mainland and K’gari time periods and between time periods on K’gari (F); c H over time – a higher number indicates a higher level of homozygosity; d Tukey test results for statistically significant differences between the mainland and K’gari time periods and between time periods on K’gari (H). (Time periods are A: 1996–2001; B: 2002–2006; C:2007–2011; D: 2012–2017; E: 2018–2020)
Within the K’gari dingo population, individual-level genetic diversity decreased over time. F values were significantly lower (less inbred) in time period A compared to time period D (Tukey test; Fig. 3b). H values were significantly lower (i.e., more diverse) in time period A compared to time periods C, D and E (Tukey test; Fig. 3d).
K’gari population - spatial structure
No genetic clustering across the island was detected by the TESS analysis (Online Resource 6). The cross-validation scores did not plateau, indicating that there was no detectable structure in the data (Online Resource 6). Furthermore, maps of K = 2 up to K = 6 did not reveal any meaningful patterns to suggest spatial clustering of genetic variation across the island (Online Resource 6). PopCluster results (described below) sorted by management areas (‘base areas’) also did not reveal any meaningful patterns (Online Resource 7).
K’gari population - genetic isolation over time
Evidence of change in the ancestral composition of K’gari dingoes between time periods was detected by both the PopCluster and PCoA analyses (Fig. 4, Online Resource 8). In the PopCluster analysis, K = 2 or 3 was supported by the DLK2 and/or FSTIS values in PopCluster (Online Resource 9). The relative ancestral proporitions between time periods were significantly different from one another (MANOVA; F(1,4) = 8.44, P = 2.8 × 10−10). Specifically, time period A was significantly different from all other time periods for cluster I (yellow) and significantly different from time periods C through E for cluster III (dark green; Fig. 4c and d).
PopCluster Admixture analysis of K’gari samples from this study in the five time periods used in other analysis (e.g. some individuals are included more than once if they were alive in more than one time period); a K = 2 through K = 4, sorted by Q value; b Box and whisker plot of relative ancestry proportions (K = 3) over time; ancestral cluster I (yellow), ancestral cluster II (pale green), ancestral cluster III (dark green); c Tukey test outcome: cluster I; d Tukey test outcome: cluster III. (A: 1996–2001; B: 2002–2006; C: 2007–2011; D: 2012–2017; E: 2018–2020)
In the PCoA, time period A was significantly different from time periods C, D and E and time period B was significantly different from time periods D and E based on the PERMANOVA (F(1,4) = 7.94, P = 0.001) and post hoc testing (Fig. 5).
K’gari population - fine scale temporal analysis of relatedness estimates
Within K’gari average relatedness (r) increased significantly over time (ANOVA: df = 4, F = 235, P < 2 × 10−16), with all time periods significantly different from one another apart from time periods B and C; D and E (Tukey test; Fig. 6a: Mean, standard error, minimum and maximum values per time period are reported in Online Resource 10). The relatedness heat map of individuals based on birth year also supports a steady increase in pairwise relatedness estimates over time: the warmer the colour, the higher the pairwise relatedness. Thus, a greater number of individuals in later time periods were more closely related (higher concentration of warmer colours) compared to earlier time periods (higher concentration of cooler colours; Fig. 6b).
Pairwise relatedness (Wang estimator) between individuals on K’gari a Box and whisker plot of individuals alive in five time periods with ANOVA and Tukey testing (inset) to show which groups were significantly different. b Relatedness heatmap with individuals in order of birth year divided into the time periods defined in Table 1; colours indicate the level of pairwise relatedness with warmer colours indicating higher pairwise relatedness. (Time periods (A: 1996–2001; B: 2002–2006; C: 2007–2011; D: 2012–2017; E: 2018–2020)
Discussion
Our study reveals key roles for both historical geographic isolation and recent management actions in determining contemporary patterns of genetic variation in K’gari dingoes. Analysis of genome-wide SNPs representing ~ 25 years of population sampling demonstrates the genetic isolation of K’gari dingoes from those on the mainland. The population is characterised by reduced genetic variation relative to mainland dingoes and likely experienced considerable drift both before and during the study period. Moreover, despite the social organization of dingoes (Corbett and Newsome 1975; Catling et al. 1992; Thomson et al. 1992), we did not find any evidence for spatial genetic clustering at the level of social groups (pack) across the island, suggesting high levels of genetic connectivity characterise the dispersal ecology of K’gari dingoes.
K’gari dingoes are, on average, more related across the population today than in the recent past and have significantly higher inbreeding coefficients than their mainland counterparts. However, limited observations in K’gari dingoes of abnormal physical and demographic characteristics, typically associated with inbreeding depression in mammals, suggests that they may have not exceeded their genomic tolerance for inbreeding. Similarly high inbreeding coefficients without associated fitness effects have been observed in California island dwarf grey foxes (Urocyon littoralis) when compared with mainland grey fox counterparts (Urocyon cinereoargenteus), where lack of inbreeding depression is associated with purging of deleterious alleles (Robinson et al. 2018). However, the effects of negative demographic influences on extinction probability of very small populations, such as the grey wolves (Canis lupus) on Isle Royale (Hedrick et al. 2019), must also be considered. Therefore, to better understand extinction probability in the K’gari dingo population, future research should focus on quantifying fertility, reproductive success and survivorship, in addition to post-mortem assessments of general condition and disease status.
K’gari dingoes - a classic example of the ‘small and isolated’ population paradigm?
Multiple lines of genetic evidence collectively support the ecological and evolutionary isolation of K’gari dingoes from their mainland counterparts, despite the island’s proximity to the mainland (this study; Cairns et al. 2018; Conroy et al. 2021). Here, both PCoA and PopCluster analysis of SNP data indicate clear differentiation of K’gari and mainland individuals (Fig. 2 and Online Resource 4), including differentiation from the geographically closest dingoes just a few kilometers away across the Great Sandy Strait (Korrawinga) at Rainbow Beach (Fig. 1). Dingoes likely arrived in the K’gari region within the last 3500–11 000 years bp; therefore, dingoes may have already established on K’gari before it was fully separated from the mainland. After K’gari’s isolation, assisted migration of dingoes with the Butchulla People, the island’s Traditional Custodians, may have also occurred (Dooley and Clode 2019). While contemporary natural immigration events and assisted (illegal) migration of individuals might theoretically occur on occasion, (Conroy et al. 2021), strong founder effects and genetic drift have likely led to differentiation of K’gari dingoes, despite the potential for connectivity with nearby mainland groups.
Given recent population size estimates (Appleby and Jones 2011; Allen et al. 2015; Conroy et al. 2017), together with low effective population size (Ne) suggested by both the species’ hierarchical social structure with alpha breeding pairs (Catling et al. 1992) and estimates from microsatellite loci (Conroy et al. 2021), we were also interested in the role of ongoing genetic drift in the population. Relative to the mainland, K’gari dingoes are characterised by both reduced SNP (this study; Fig. 3c) and microsatellite (Conroy et al. 2021) genetic variation. That an island population supports reduced variation is not unexpected (Frankham 1996, 1997). Repeated periods of intensified genetic drift during the original arrival(s) of dingoes to Australia, subsequent local founder events, and anecdotal population declines via 19th century baiting/culling events, all likely contribute to both broad and fine-scale patterns of variation in contemporary populations (Petrie 1995; Cairns et al. 2018, 2023; Conroy et al. 2021; Stephens et al. 2022; Kumar et al. 2023). Observed changes in ancestry proportions i.e. the presence of ancestral genetic clusters in the data on K’gari over the period of our study (Fig. 4.), also suggests that as an ecologically closed population it remains highly susceptible to the chance effects of drift. Ancestry proportions varied over the sampling periods demonstrating a pattern of sequential change over the past ~ 25 years (K = 3; Fig. 4). Demographic parameters that influence Ne, like population size and mating system, are important in determining the strength of genetic drift in small populations (Allendorf et al. 2013). However, drift can also outweigh the relative strength of selection in small populations, reducing its efficacy when selection is weak (Wright 1931); under this scenario, increased loss and fixation of alleles via drift may have important consequences for adaptive potential in an island population like K’gari (Bijlsma and Loeschcke 2012; James et al. 2016; Willi et al. 2022). Indeed, Kumar et al. (2023) argue that despite their ecological success across Australia, mainland dingoes themselves may carry a substantial genomic load from repeated bottlenecks and founder effects; dingoes sustain a larger deleterious mutational load than breed dogs, village dogs and wolves, and have the lowest genomic diversity of all canids studied (Freedman et al. 2014; Zhang et al. 2020; Kumar et al. 2023). A well-sampled genomic perspective of K’gari dingoes would be a valuable addition to existing studies and has the potential to provide informative genetic markers associated with fitness for future monitoring.
Both mean pairwise relatedness (Fig. 6, Online Resource 10) and mean inbreeding (Fig. 3, Online Resource 5) in Kgari’s dingoes are characteristic of a population experiencing minimal immigration of genetically divergent individuals. Mean pairwise relatedness within the population has increased over time (Fig. 6a, Online Resource 10), and this trend is very clear in our heat map analysis of individuals (Fig. 6b); based on birth year, K’gari dingoes are, on average, more closely related across the population today than in the recent past, suggesting ongoing, albeit gradual, erosion of genetic diversity. While mean inbreeding has not changed significantly over time, the population is characterised by moderate-high inbreeding values for the entirety of our study period relative to the mainland values (Fig. 3a, Online Resource 5). Together, these population genetic parameters are informative risk factors for the probability of inbreeding depression in naturally outbred populations (Hedrick and Garcia-Dorado 2016); in the absence of genetically variable migrants, K’gari’s dingoes are likely at risk of inbreeding depression. Inbreeding depression in canids can present physically as reductions in adult body size and as direct fitness declines in reproductive success, and adult and pup survival (e.g. red wolf Canis rufus (Brzeski et al. 2014), arctic fox Vulpes lagopus (Norén et al. 2016)). While no unusual physical abnormalities have been observed in Kgari’s dingoes (L Behrendorff pers. obs.), suggesting no substantive evidence of inbreeding depression overall, we have not tested here whether variation in individual inbreeding levels is correlated with variation in fitness traits. While data on some indicators of fitness such as lifespan (Behrendorff and Allen 2016) and litter sizes (Allen et al. 2015) are available, the lack of long-term pedigree data represents a key knowledge gap to understanding how inbreeding may impact the population.
While social grouping can structure genetic variation in space (Chesser 1991a, b; Parreira and Chikhi 2015), we found K’gari dingoes did not exhibit any detectable spatial genetic structure across the island, neither at a fine scale that could be representative of social groups nor at a spatially broader scale, consistent with Conroy et al. (2021). Using PCoA together with both spatially informed and non-spatial ancestry estimation, we did not detect any signatures of genetic structure across the island (Fig. 2, Online Resource 6). The most likely explanation for our finding is that ecological and genetic connectivity is sufficient to prevent discrete spatial structure of the island’s gene pool. As a result, inter-pack breeding appears common enough that packs cannot be distinguished genetically across the island. While most dingoes maintain a core stable home range, they do still have substantial capacity for movement. On any one day, individuals can travel 5–20 km whilst foraging, and dispersal beyond natal pack territories by both males and females often exceeds 100 km (and sometimes 500 km) on the mainland (Thomson et al. 1992; Allen 2009; McNeill et al. 2016). Limited by the confines of the island, most K’gari dingoes spend their time within discrete pack boundaries (Allen et al. 2015), but frequent foraging and dispersal movements at sexual maturity across the island are not uncommon for a substantial proportion of animals (Baxter and Davies 2018; Behrendorff et al. 2018; White 2021). Ear-tagged and individually recognisable dingoes are often seen outside their natal home range (L Behrendorff pers. obs.). In the absence of new migrants, and despite the presence of discrete social territories, the mating behaviour and spatial ecology of dingoes on the island promotes persistent genetic mixing of social units. This potentially influences the rate of genetic erosion in the population, delaying its entry into the classic ‘small population paradigm’ of inbreeding and population decline (Caughley 1994). While it is possible that the opportunistic sampling of individuals in our study might have influenced the detectability of social groups within our data to some extent, i.e. we may have under sampled related individuals, life history traits, especially those affecting population growth such as longer and overlapping generation times in age-structured species like dingoes, are known to buffer the effects of drift on genetic diversity (Ellner and Hairston 1994; Bishop et al. 2009). On K’gari, longevity of breeders and the frequent movement of both males and females across the island may be important mechanisms by which spatial differentiation and loss of variation via drift is limited.
Effects of past population management on contemporary genetic variation in K’gari dingoes
While contemporary management on K’gari aims to balance the ecological roles, and cultural and conservation aspects of the island’s dingoes with those of tourism, while also minimising human-wildlife conflict (Allen et al. 2015; Tapply 2018), past population management was less focused on population genetic targets. Undoubtedly, the deeper history of the population, characterised by bottlenecks and founder effects, contributes to reduced variation on K’gari today relative to mainland populations. Furthermore, even though K’gari dingoes are recognised as ‘iconic’ and culturally important through their deep-time associations with the Butchulla people (Daniels and Corbett 2003; Smith and Litchfield 2009), they have a long history of demographic flux. Reports suggest that dingoes were sometimes killed (and eaten) prior to European presence on the island (Eastwood 2012). The population experienced repeated offtake both before and after the island was gazetted as Great Sandy National Park in 1971 and achieved World Heritage status in 1992. Population reductions include offtake from the culling ‘bounty system’ in the 19th and early 20th century as a way of limiting sheep and cattle depredation (Anon 1902; Petrie 1995), and present-day selective removal of dangerous individuals as part of an adaptive management strategy (Tapply 2018; Brink et al. 2019; Behrendorff 2021). We were interested in whether the introduction of lethal management interventions on the population from 2001 had left signatures in the broader genetic composition of K’gari dingoes; noting that the population was already characterised by reduced variation and higher mean relatedness relative to the mainland at the beginning of our study period prior to these management actions (period A, Fig. 3). Between 2001 and 2013 a total of 110 dingoes were humanely destroyed (average of 6.6 dingoes per year; Allen et al. 2015), with 31 individuals killed in 2001. Relative to period A (1996–2001) we observed significant differences in estimates of homozygosity, inbreeding, and relatedness in subsequent time periods (Figs. 3 and 5). Although the demographic offtake of K’gari dingoes, skewed towards young males (Allen et al. 2015), may have been countered by compensatory increases in recruitment and survival (Frank and Woodroffe 2001), the killing of such a proportionately large number of individuals in the early 2000s nevertheless produced a detectable decrease in overall genetic diversity in the population. Given the isolation and limited effective population size (Ne) of K’gari dingoes relative to mainland populations (Conroy et al. 2021), the initial observed loss of genetic diversity is not altogether unexpected. Fortunately, this loss appears to have slowed down through time (Figs. 3 and 5) despite ongoing removals of small numbers of animals that still occur in response to dingo aggression towards people (Behrendorff 2021).
Conclusions
Contemporary patterns of genetic variation in K’gari dingoes are most likely explained by several interacting factors including small population size, isolation, and past management actions, together with historic population events such as bottlenecks and founder effects. At the starting point of our study genetic variation in the population was already significantly reduced relative to the mainland, and while management actions in the early 2000s likely reduced diversity at that time, continuing loss appears to have since slowed down; accordingly, ongoing genetic monitoring is recommended. While the population is characterised by (a) high inbreeding and reduced diversity relative to the mainland, and (b) increasing relatedness over the time periods analysed here, we hypothesise that the social structure and dispersal ecology of the species might provide a mechanism for widespread genetic connectivity across the island that buffers the intensity with which genetic drift might act on a similar population. This may delay the predicted effects of inbreeding in the population, however, such effects are inevitable in the ongoing absence of migrants.
Our findings should be considered in any management planning that incorporates measurable genetic targets set over ecologically meaningful timeframes in this population. Uninhibited movement of dingoes around the island is important for minimising the effects of both localised inbreeding and genetic drift and needs to be maintained. If maintaining or increasing genetic diversity of K’gari dingoes is identified as a management priority, intentional immigration is an option; however, strategies to minimise negative human-dingo interactions, and thereby lethal management interventions, also remain important. The social ecology of dingoes is a key contributor to their ecological success and persistence. Moving forward, a more detailed understanding of the island’s dispersal dynamics will provide a valuable scaffold for interpreting population genetic and genomic metrics within an associated management framework.
Data availability
All data used in this manuscript are archived at zivahub.uct.ac.za doi: 10.25375/uct.24798171.v1.
References
Abdellaoui A, Hottenga JJ, Knijff P, De et al (2013) Population structure, migration, and diversifying selection in the Netherlands. Eur J Hum Genet 21:1277–1285. https://doi.org/10.1038/ejhg.2013.48
Allen LR (2009) Best practice baiting: dispersal and seasonal movement of wild dogs (Canis lupus familiaris) In: Technical highlights. Invasive plant and animal research 2008-09. Brisbane, pp 61–62
Allen BL, Higginbottom K, Bracks JH et al (2015) Balancing dingo conservation with human safety on Fraser Island: the numerical and demographic effects of humane destruction of dingoes. Australas J Environ Manag 22:197–215. https://doi.org/10.1080/14486563.2014.999134
Allendorf FW, Luikart G, Aitken SN (2013) Conservation and the genetics of populations, 2nd edn. Wiley Blackwell, Oxford, UK
Anon (1902) General News. Maryborough Chronicle, Wide Bay and Burnett Advertiser 19Dec1902:2. https://doi.org/https://trove.nla.gov.au/newspaper/article/148183960
Appleby R, Jones D (2011) Analysis of Preliminary Dingo Capture-Mark-Recapture Experiment on Fraser Island: final Report to Queensland Parks and Wildlife Service. Brisbane
Arauco-Shapiro G, Schumacher KI, Boersma D, Bouzat JL (2020) The role of demographic history and selection in shaping genetic diversity of the Galápagos penguin (Spheniscus mendiculus). PLoS ONE 15:e0226439. https://doi.org/10.1371/journal.pone.0226439
Arbizu PM (2017) pairwiseAdonis: Pairwise Multilevel Comparison using Adonis. R package version 0.4
Archer-Lean C, Wardell-Johnson A, Conroy G, Carter J (2015) Representations of the dingo: contextualising iconicity. Australas J Environ Manag 22:181–196. https://doi.org/10.1080/14486563.2014.985268
Ballard JWO, Field MA, Edwards RJ et al (2023) The Australasian dingo archetype: de novo chromosome-length genome assembly, DNA methylome, and cranial morphology. Gigascience 12:giad018. https://doi.org/10.1093/GIGASCIENCE/GIAD018
Balme J, Connor SO, Fallon S (2018) New dates on dingo bones from Madura Cave provide oldest firm evidence for arrival of the species in Australia. Sci Rep 8:9933. https://doi.org/10.1038/s41598-018-28324-x
Baxter G, Davies N (2018) Movements of dingoes on K’gari-Fraser Island: implications for management. Australas J Environ Manag 25:132–146. https://doi.org/10.1080/14486563.2017.1397062
Behrendorff L (2021) Best-practice dingo management: six lessons from K’gari (Fraser Island). Australian Zoologist 41:521–533. https://doi.org/10.7882/AZ.2021.001
Behrendorff L, Allen BL (2016) From den to dust: longevity of three dingoes (Canis lupus dingo) on Fraser Island (K’gari). Aust Mammal 38:256–260. https://doi.org/10.1071/AM16005
Behrendorff L, Leung LKP, Allen BL (2018) Utilisation of stranded marine fauna washed ashore on K’gari (Fraser Island), Australia, by dingoes. Aust J Zool 66:128–138. https://doi.org/10.1071/ZO18022
Bergström A, Frantz L, Schmidt R et al (2020) Origins and genetic legacy of prehistoric dogs. Science 370:557–564. https://doi.org/10.1126/science.aba9572
Bijlsma R, Loeschcke V (2012) Genetic erosion impedes adaptive responses to stressful environments. Evol Appl 5:117–129. https://doi.org/10.1111/j.1752-4571.2011.00214.x
Bishop JM, Leslie AJ, Bourquin SL, O’Ryan C (2009) Reduced effective population size in an overexploited population of the Nile crocodile (Crocodylus niloticus). Biol Conserv 142:2335–2341. https://doi.org/10.1016/j.biocon.2009.05.016
Bouzat JL (2010) Conservation genetics of population bottlenecks: the role of chance, selection, and history. Conserv Genet 11:463–478. https://doi.org/10.1007/s10592-010-0049-0
Brink HA, Purcell BV, Letnic M et al (2019) Pets and pests: a review of the contrasting economics and fortunes of dingoes and domestic dogs in Australia, and a proposed new funding scheme for non-lethal dingo management. Wildl Res 46:365–377. https://doi.org/10.1071/WR19030
Brook BW, Tonkyn DW, O’Grady JJ, Frankham R (2002) Contribution of inbreeding to extinction risk in threatened species. Conserv Ecol 6:16. https://www.jstor.org/stable/2386988
Brzeski KE, Rabon DR, Chamberlain MJ et al (2014) Inbreeding and inbreeding depression in endangered red wolves (Canis rufus). Mol Ecol 23:4241–4255. https://doi.org/10.1111/MEC.12871
Cairns KM (2021) What is a dingo – origins, hybridisation and identity. Aust Zool 41:322–337. https://doi.org/10.7882/AZ.2021.004
Cairns KM, Wilton AN (2016) New insights on the history of canids in Oceania based on mitochondrial and nuclear data. Genetica 144:553–565. https://doi.org/10.1007/s10709-016-9924-z
Cairns KM, Brown SK, Sacks BN, Ballard JWO (2017) Conservation implications for dingoes from the maternal and paternal genome: multiple populations, dog introgression, and demography. Ecol Evol 7:9787–9807. https://doi.org/10.1002/ece3.3487
Cairns KM, Shannon LM, Koler-Matznick J et al (2018) Elucidating biogeographical patterns in Australian native canids using genome wide SNPs. PLoS ONE 13:e0198754. https://doi.org/10.1371/journal.pone.0198754
Cairns KM, Crowther MS, Nesbitt B, Letnic M (2022) The myth of wild dogs in Australia: are there any out there? Aust Mammal 44:67–75. https://doi.org/10.1071/AM20055
Cairns KM, Crowther MS, Parker HG et al (2023) Genome-wide variant analyses reveal new patterns of admixture and population structure in Australian dingoes. Mol Ecol 32:4133–4150. https://doi.org/10.1111/mec.16998
Carter J, Wardell-Johnson A, Archer-Lean C (2017) Butchulla perspectives on dingo displacement and agency at K’gari-Fraser Island, Australia. Geoforum 85:197–205. https://doi.org/10.1016/j.geoforum.2017.08.001
Catling PC, Corbett LK, Newsome AE (1992) Reproduction in captive and wild dingoes (Canis familiaris Dingo) in temperate and arid environments of Australia. Wildl Res 19:195–209. https://doi.org/10.1071/WR9920195
Caughley G (1994) Directions in conservation biology. J Anim Ecol 63:215–244. https://doi.org/10.2307/5542
Caye K, Francois O (2016) tess3r: inference of spatial population genetic structure, R package version 1.1.0
Caye K, Deist TM, Martins H et al (2016) TESS3: fast inference of spatial population structure and genome scans for selection. Mol Ecol Resour 16:540–548. https://doi.org/10.1111/1755-0998.12471
Chesser RK (1991a) Influence of gene flow and breeding tactics on gene diversity within populations. Genetics 129:573–583. https://doi.org/10.1093/genetics/129.2.573
Chesser RK (1991b) Gene Diversity and Female Philopatry. Genetics 127:437–447. https://doi.org/10.1093/genetics/129.2.573
Conroy GC, Ogbourne SM, Lamont RW, Wardell-Johnson A, Bridges L (2017) A baseline genetic analysis of the K’gari-Fraser Island dingo population. Univ Sunshine Coast https://research.usc.edu.au/esploro/outputs/99450869302621
Conroy GC, Lamont RW, Bridges L, Stephens D, Wardell-Johnson A, Ogbourne SM (2021) Conservation concerns associated with low genetic diversity for K’gari–Fraser Island dingoes. Sci Rep 11:9503. https://doi.org/10.1038/s41598-021-89056-z
Corbett LK (1988) Social Dynamics of a Captive Dingo Pack: Population Regulation by Dominant Female Infanticide. Ethology 78:177–198. https://doi.org/10.1111/J.1439-0310.1988.TB00229.X
Corbett L (2001) The dingo in Australia and Asia, 2nd edn. JB Books, West Torrens, SA, Australia
Corbett LK, Newsome AE (1975) Dingo society and its maintenance: a preliminary analysis. In: Fox M (ed) Wild canids: their systematics, behavioural ecology and evolution. Van Nostrand Reinhold, New York, pp 369–379
Daniels MJ, Corbett L (2003) Redefining introgressed protected mammals: when is a wildcat a wild cat and a dingo a wild dog? Wildl Res 30:213–218. https://doi.org/10.1071/WR02045
Donfrancesco V, Allen BL, Appleby R et al (2023) Understanding conflict among experts working on controversial species: a case study on the Australian dingo. Conserv Sci Pract 5:e12900. https://doi.org/10.1111/csp2.12900
Dooley G, Clode D (2019) The First Wave: exploring early coastal contact history in Australia. Wakefield, Adelaide
Eastwood K (2012) Finding Fraser Island. Australian Geographic Magazine 107:65–79
Elgar MA, Clode D (2001) Inbreeding and extinction in Island populations: a cautionary note. Conserv Biol 15:284–286. https://doi.org/https://www.jstor.org/stable/2641670
Ellner S, Hairston NG (1994) Role of overlapping generations in maintaining genetic variation in a fluctuating environment. Am Nat 143:403–417. https://www.jstor.org/stable/2462737
Frank L, Woodroffe R (2001) Behaviour of carnivores in exploited and controlled populations. In: Gittleman S, Funk S, Macdonald D, Wayne R (eds) Carnivore Conservation. Cambridge University Press, Cambridge, pp 419–442
Frankham R (1995) Inbreeding and extinction: a threshold effect. Conserv Biol 9:792–799. https://doi.org/10.1046/J.1523-1739.1995.09040792.X
Frankham R (1996) Relationship of genetic variation to Population size in Wildlife. Conserv Biol 10:1500–1508. https://doi.org/10.1046/j.1523-1739.1996.10061500.x
Frankham R (1997) Do island populations have less genetic variation than mainland populations? Heredity 78:311–327. https://doi.org/10.1038/hdy.1997.46
Frankham R (1998) Inbreeding and extinction: island populations. Conserv Biol 12:665–675. https://doi.org/10.1111/J.1523-1739.1998.96456.X
Frankham R (2005) Genetics and extinction. Biol Conserv 126:131–140. https://doi.org/10.1016/j.biocon.2005.05.002
Frankham R, Ralls K (1998) Inbreeding leads to extinction. Nature 392:441–442. https://doi.org/10.1038/33022
Freedman AH, Gronau I, Schweizer RM et al (2014) Genome sequencing highlights the dynamic early history of Dogs. PLoS Genet 10:1004016. https://doi.org/10.1371/journal.pgen.1004016
Gontz AM, Moss PT, Sloss CR et al (2015) Understanding past climate variation and environmental change for the future of an iconic landscape – K’gari Fraser Island, Queensland, Australia. Australas J Environ Manag 22:105–123. https://doi.org/10.1080/14486563.2014.1002120
Gooley R, Hogg CJ, Belov K, Grueber CE (2017) No evidence of inbreeding depression in a tasmanian devil insurance population despite significant variation in inbreeding. Sci Rep 7:1830. https://doi.org/10.1038/s41598-017-02000-y
Hedrick PW (1999) Highly variable loci and their interpretation in evolution and conservation. Evol (N Y) 53:313–318. https://doi.org/10.2307/2640768
Hedrick PW, Garcia-Dorado A (2016) Understanding Inbreeding Depression, purging, and genetic rescue. Trends Ecol Evol 31:940–952. https://doi.org/10.1016/J.TREE.2016.09.005
Hedrick PW, Robinson JA, Peterson RO, Vucetich JA (2019) Genetics and extinction and the example of Isle Royale wolves. Anim Conserv 22:302–309. https://doi.org/10.1111/acv.12479
Jackson SM, Fleming PJS, Eldridge MDB et al (2021) Taxonomy of the Dingo: it’s an ancient dog. Aust Zool 41:347–357. https://doi.org/10.7882/AZ.2020.049
James JE, Lanfear R, Eyre-walker A (2016) GBE Molecular Evolutionary consequences of Island colonization. Genome Biol Evol 8:1876–1888. https://doi.org/10.1093/gbe/evw120
Kumar M, Conroy G, Ogbourne S et al (2023) Genomic signatures of bottleneck and founder effects in dingoes. Ecol Evol 13:e10525. https://doi.org/10.1002/ECE3.10525
Leach D, Shaw AK, Weiss-Lehman C (2020) Stochasticity in social structure and mating system drive extinction risk. Ecosphere 11:e03038. https://doi.org/10.1002/ecs2.3038
Letnic M, Ritchie EG, Dickman CR (2012) Top predators as biodiversity regulators: the dingo Canis lupus dingo as a case study. Biol Rev 87:390–413. https://doi.org/10.1111/J.1469-185X.2011.00203.X
Lindblad-Toh K, Wade CM, Mikkelsen TS et al (2005) Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nat 2005 438:803–819. https://doi.org/10.1038/nature04338
Lippe C, Dumont P, Bernatchez L et al (2006) High genetic diversity and no inbreeding in the endangered copper redhorse, Moxostoma hubbsi (Catostomidae, Pisces): the positive sides of a long generation time. Mol Ecol 15:1769–1780. https://doi.org/10.1111/j.1365-294X.2006.02902.x
Lord K, Feinstein M, Smith B, Coppinger R (2013) Variation in reproductive traits of members of the genus Canis with special attention to the domestic dog (Canis familiaris). Behav Process 92:131–142. https://doi.org/10.1016/j.beproc.2012.10.009
MacArthur RH, Wilson EO (1963) An equilibrium theory of Insular Zoogeography. Evol (N Y) 17:373. https://doi.org/10.2307/2407089
Marees AT, de Kluiver H, Stringer S et al (2018) A tutorial on conducting genome-wide association studies: quality control and statistical analysis. Int J Methods Psychiatr Res 27:e1608. https://doi.org/10.1002/mpr.1608
McNeill AT, Leung LK, Goullet MS et al (2016) Dingoes at the Doorstep: Home Range sizes and activity patterns of dingoes and other Wild Dogs around Urban areas of North-Eastern Australia. Animals 6:48. https://doi.org/10.3390/ani6080048
Meiring C, Schurz H, van Helden P et al (2022) African wild dogs (Lycaon pictus) from the Kruger National Park, South Africa are currently not inbred but have low genomic diversity. Sci Rep 12:14979. https://doi.org/10.1038/s41598-022-19025-7
Meszaros G (2021) Genomics Boot Camp. https://genomicsbootcamp.github.io/book/. Accessed 22 Apr 2022
Norén K, Godoy E, Dalén L et al (2016) Inbreeding depression in a critically endangered Carnivore. Mol Ecol 25:3309–3318. https://doi.org/10.1111/mec.13674
Oksanen J, Blanchet FG, Friendly M et al (2020) vegan: Community Ecology R package, version 2.6-4
Ørsted M, Hoffmann AA, Sverrisdóttir E et al (2019) Genomic variation predicts adaptive evolutionary responses better than population bottleneck history. PLoS Genet 15:e1008205. https://doi.org/10.1371/journal.pgen.1008205
Parreira BR, Chikhi L (2015) On some genetic consequences of social structure, mating systems, dispersal, and sampling. Proc Natl Acad Sci U S A 112:E3318–E3326. https://doi.org/10.1073/pnas.1414463112
Petrie R (1995) Early days on Fraser Island 1913–1922, 1st edn. Go Bush Safaris, Gladesville, NSW, Australia
Pimm SL (1991) The balance of nature? Ecological issues in the conservation of species and communities. Chicago University Press, Chicago
Purcell S (2022) PLINK v1.9 http://pngu.mgh.harvard.edu/purcell/plink/
Purcell S, Neale B, Todd-Brown K et al (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
R Core Team (2022) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Robinson JA, Brown C, Kim BY et al (2018) Purging of strongly deleterious mutations explains long-term persistence and absence of Inbreeding Depression in Island Foxes. Curr Biol 28:3487–3494e4. https://doi.org/10.1016/J.CUB.2018.08.066
RStudio Team (2022) RStudio: Integrated Development Environment for R. PBC, Boston, MA. http://www.rstudio.com/
Smith BP (2015) The dingo debate: origins, behaviour, and conservation. CSIRO Publishing ISBN, p 9781486300303
Smith BP, Litchfield CA (2009) A review of the relationship between indigenous australians, dingoes (Canis dingo) and domestic dogs (Canis familiaris). Anthrozoos 22:111–128. https://doi.org/10.2752/175303709X434149
Smith BP, Cairns KM, Adams JW et al (2019) Taxonomic status of the Australian dingo: the case for Canis dingo Meyer, 1793. Zootaxa 4564:173–197. https://doi.org/10.11646/zootaxa.4564.1.6
Stephens D, Wilton AN, Fleming PJS, Berry O (2015) Death by sex in an Australian icon: a continent-wide survey reveals extensive hybridization between dingoes and domestic dogs. Mol Ecol 24:5643–5656. https://doi.org/10.1111/mec.13416
Stephens D, Fleming PJS, Sawyers E, Mayr TP (2022) An isolated population reveals greater genetic structuring of the Australian dingo. Sci Rep 12:1–12. https://doi.org/10.1038/s41598-022-23648-1
Tapply J (2018) Contemporary dingo management on K’gari (Fraser Island, Great Sandy National Park) under the Queensland Parks and Wildlife Service. Australas J Environ Manag 25:119–131. https://doi.org/10.1080/14486563.2017.1422042
Thomson PC, Rose K, Kok NE (1992) The behavioural ecology of dingoes in north-western Australia. VI. Temporary extraterritorial movements and dispersal. Wildl Res 19:585–595. https://doi.org/10.1071/WR9920585
Torres-Florez JP, Hucke-Gaete R, Rosenbaum H, Figueroa CC (2014) High genetic diversity in a small population: the case of Chilean blue whales. Ecol Evol 4:1398–1412. https://doi.org/10.1002/ece3.998
Vellend M, Geber MA (2005) Connections between species diversity and genetic diversity. Ecol Lett 8:767–781. https://doi.org/10.1111/J.1461-0248.2005.00775.X
Walker KE, Baldwin C, Conroy GC et al (2022) Ecological and cultural understanding as a basis for management of a globally significant Island Landscape. Coasts 2:152–202. https://doi.org/10.3390/COASTS2030009
Wang J (2011) COANCESTRY: a program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol Ecol Resour 11:141–145. https://doi.org/10.1111/j.1755-0998.2010.02885.x
Wang J (2022) Fast and accurate population admixture inference from genotype data from a few microsatellites to millions of SNPs. Heredity 129:79–92. https://doi.org/10.1038/s41437-022-00535-z
White NJ (2021) Spatio-temporal ecology of dangerous wildlife in a conservation setting, and implications for management of human-wildlife interactions: wongari (dingoes -. Canis dingo) on K’gari (Fraser Island), Queensland, Australia. The University of Queensland, PhD Thesis
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer-, New York
Wickham H, Averick M, Bryan J et al (2019) Welcome to the Tidyverse. J Open Source Softw 4:1686. https://doi.org/10.21105/JOSS.01686
Willi Y, Kristensen TN, Sgro CM et al (2022) Conservation genetics as a management tool: the five best-supported paradigms to assist the management of threatened species. PNAS 119:e2105076119. https://doi.org/10.1073/pnas.2105076119
Wright S (1931) Evolution in mendelian populations. Genetics 16:290. https://doi.org/10.1093/genetics/16.2.97
Zhang S, Wang GD, Ma P et al (2020) Genomic regions under selection in the feralization of the dingoes. Nat Commun 11:671. https://doi.org/10.1038/s41467-020-14515-6
Acknowledgements
The authors acknowledge the Butchulla People as the traditional custodians of K’gari.
Funding
Open access funding provided by University of Cape Town. This project was funded by the Queensland Government Department of Environment and Science, via a grant to BLA and JMB (USQ631472014).
Author information
Authors and Affiliations
Contributions
LB and BA conceptualised the study. BA secured the funding and invited all other contributions from stakeholders and conservation genetics experts. LB, RLA, GB, JWOB, KMC, GCC, PJSF, CEG, JO, DAS, DS, CW, JMB and SM were involved in developing the analytical framework for the study. LB provided samples for SNP genotyping and associated metadata for the analysis. SMM performed the analysis and prepared the figures and tables with assistance from JMB. All authors were involved in the interpretation of the results. SMM and JMB wrote the manuscript. All authors provided feedback on the initial drafts and approved the final submitted version.
Corresponding author
Ethics declarations
Competing interests
KMC is a scientific advisor to the Australian Dingo Foundation and the New Guinea Highland Wild Dog Foundation. KMC (co-chair), BLA (co-chair), LB, and GAB are members of the IUCN Canid Specialist Group Dingo Working Group.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Miller, S.M., Behrendorff, L., Allen, B.L. et al. Isolation, small population size, and management influence inbreeding and reduced genetic variation in K’gari dingoes. Conserv Genet (2024). https://doi.org/10.1007/s10592-024-01616-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10592-024-01616-8