Abstract
Small populations of endangered species risk losing already eroded genetic diversity, important for adaptive potential, through the effects of genetic drift. The magnitude of drift can be mitigated by maximising the effective population size, as is the goal of genetic management strategies. Different mating systems, specifically those leading to reproductive skew, exacerbate genetic drift by distorting contributions. In the absence of an active management strategy, reproductive skew will have long-term effects on the genetic composition of a population, particularly where admixture is present. Here we examine the contrasting effects of conservation management strategies in two ex situ populations of the Critically Endangered eastern black rhino (Diceros bicornis michaeli), one managed as a semi-wild population in South Africa (SAx), and one managed under a mean-kinship breeding strategy in European zoos. We use molecular data to reconstruct pedigrees for both populations and validate the method using the zoo studbook. Using the reconstructed pedigree and studbook we show there is male sex-specific skew in both populations. However, the zoo’s mean-kinship breeding strategy effectively reduces reproductive skew in comparison to a semi-wild population with little genetic management. We also show that strong male reproductive skew in SAx has resulted in extensive admixture, which may require a re-evaluation of the population’s original intended role in the black rhino meta-population. With a high potential for admixture in many ex situ populations of endangered species, molecular and pedigree data remain vital tools for populations needing to balance drift and selection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the face of genetic erosion through defaunation (Bogoni et al. 2020), and range contraction (Britnell et al. 2023; Pacifici et al. 2020), ex situ populations of endangered species may serve as valuable reservoirs of genetic diversity and individuals for reintroduction (Farré et al. 2022). These, typically small, populations are vulnerable to inherent risks, including low resilience to perturbation and stochastic effects, and risk being driven into an extinction vortex through genetic drift and inbreeding depression (Williams et al. 2021). Genetic management of ex situ populations to minimize drift and preserve valuable diversity is therefore a major challenge for endangered species conservation and the restoration of resilient populations (DeWoody et al. 2021; García-Dorado & Caballero 2021; Hohenlohe et al. 2021).
One approach to minimize the effects of drift in ex situ populations is to employ active management strategies to reduce reproductive skew (Gooley et al. 2018). These may include male rotation strategies for group-living animals (Leus et al. 2011; Mucha & Komen 2016) or direct interventions to bypass competition, such as artificial insemination (Howard et al. 2016). In captive breeding programmes, current best practice to minimize drift is to equalise individual genetic contributions using a mean-kinship breeding strategy informed by a multi-generational pedigree. Under this strategy individuals are paired to minimize the average relatedness within the population (Ivy & Lacy 2012; Willoughby et al. 2015). An alternative management approach is to maintain animals in semi-wild conditions with natural mate choice. Doing so has the advantage of maintaining selection pressure to retain adaptive genetic variation, but at the potential expense of loss of genetic diversity due to high variance in mating success (Cain et al. 2014). Another factor that can impact the value of ex situ populations for reintroduction is if they have been founded or supplemented using animals of diverse or unknown origins, whether opportunistically or intentionally (Modesto et al. 2018; Senn et al. 2019). In this study we evaluate the conservation value and reintroduction potential of two ex situ populations of the Critically Endangered black rhino (Diceros bicornis, Emslie 2020) that exemplify the management strategies and challenges outlined above.
The black rhino has undergone a precipitous bottleneck from over 850,000 animals to only ~ 6000 today due to the poaching crisis (Moodley et al. 2017), and the ongoing threat of poaching necessitates the protection of a network of small and isolated populations (Emslie 2020). Ex situ populations are a key component of the meta-population, serving as sources of animals for translocations. But while small populations are easier to protect and manage, they suffer the compounding costs of genetic drift and inbreeding which are amplified with decreasing population size. Due to the severe bottleneck, the black rhino has suffered substantial genetic erosion (Moodley et al. 2017), so minimising further genetic drift in the extant small populations is important to maintain what little diversity remains. Reproductive skew has been identified as a priority concern for the species (Kenya Wildlife Service 2022). Female reproductive skew has been demonstrated among black rhino in both captive (Edwards et al. 2015a, b) and free-ranging populations, with variation in lifetime reproductive success (Harvey Sky et al. 2022), inter-calving interval (Patton et al. 2008) and age of first reproduction (Law et al. 2013). Male reproductive skew has been found to be even more extreme among freely mixing black rhino in semi-wild conditions (Garnier et al. 2001) with dominant males securing the highest number of matings with females (Cain et al. 2014).
Previous management of the species as four ‘ecotypes’, that became associated with historic subspecies classifications (eastern D. b. michaeli, southern D. b. minor, south-western D. b. bicornis and western D. b. longipes, Rookmaaker 1995), has left a high potential for admixture in ex situ populations. Based on recent genomic analyses, the eastern black rhino is considered to comprise multiple evolutionary significant units (ESUs, Sánchez-Barreiro et al. 2023), which have likely been mixed both in situ (Muya et al. 2011) and ex situ. The southern ESU (now reclassified as D. b. bicornis, Sánchez-Barreiro et al. 2023) is considered less diverse, but shows significant divergence from the eastern clade, estimated to have diverged 641Ka (Moodley et al. 2020). These two deepest clades are divided by the Zambezi river which serves as a significant barrier to animal movement, although rare instances of admixture along this boundary are still possible (Sánchez-Barreiro et al. 2023). Admixture has the potential to release the burden of inbreeding depression, but with increasing divergence may result in outbreeding depression (Frankham et al. 2011). Where admixture and reproductive skew occur simultaneously, the introgressed alleles may be eliminated, or conversely, may spread disproportionately throughout the population. In its most extreme form this results in a genetic sweep and the erosion of ‘native’ alleles (Adams et al. 2011; Gibson et al. 2019). Therefore, reproductive skew not only accelerates genetic drift but can dramatically alter the evolutionary trajectory of a population in the context of admixture.
Pedigrees form a critical management tool for the black rhino artificial meta-population, as it is almost entirely reliant on human-mediated translocations to maintain gene flow (Emslie 2020). Accurately constructing and maintaining a multi-generational pedigree requires intense sampling and long-term monitoring of a population (Galla et al. 2022), which is challenging for free-ranging animals. Furthermore, the accuracy of primary assumptions dictates the subsequent accuracy of a pedigree. For example, founders are often assumed to be unrelated which, if untrue, may result in highly inbred populations with negative long-term implications for breeding programmes (Hogg et al. 2019). Conversely, if founders are incorrectly assumed to belong to the same evolutionary group, this can result in genetic admixture in populations of conservation concern which may compromise their conservation value (Hvilsom et al. 2013). By combining observational data with molecular information, many of the assumptions and pitfalls of an incomplete pedigree can be addressed. Molecular data has been used to correct studbook records for captive African painted dogs (Lycaon pictus, Miller-Butterworth et al. 2021), captive and reintroduced Californian condors (Gymnogyps californianus, Moran et al. 2021), and has revealed relatedness amongst founders in populations of the Critically Endangered Kākāpo (Strigops habroptilus, Bergner et al. 2014).
In this study we incorporate molecular data with studbook records for the management of two key ex situ populations of the Critically Endangered eastern black rhino: the population of eastern black rhino in European Zoos managed under the European Association of Zoos and Aquaria (EAZA) Endangered Species Programme (EEP), and an ex situ population of eastern black rhino that is extralimital in South Africa (SAx). Admixture with the southern black rhino is known to be present in the SAx population, although the full extent of this has not been established (Kretzschmar, pers. comm.). Understanding admixture in conservation populations is key to inform both conservation and management strategy, and will dictate the suitability of individual animals for different reintroduction plans.
Here we use the genotypes obtained from a panel of 15 autosomal microsatellite loci combined with mitochondrial d-loop haplotypes to: 1) validate the marker panel for pedigree reconstruction using the studbook of European zoos and reconstruct the pedigree of the SAx population of admixed black rhino; 2) compare the genetic diversity maintained in the two ex situ populations under different management strategies (mean-kinship breeding in the European zoos and free-ranging in SAx); 3) evaluate the effectiveness of the management strategies at moderating reproductive skew and therefore genetic drift; and, 4) quantify the contribution of southern ancestry to the zoo and SAx populations. Based on these analyses we discuss the implications for conservation strategies utilizing animals from these two major ex situ populations of eastern black rhino.
Methods
Ethical approval for sampling of European zoo animals was granted by the EEP Taxon Advisory Group for rhino. Ethical approval for the use of veterinary by-product samples from South Africa was granted by the University of Manchester, UK. Tissue samples from SAx were imported from South Africa under CITES export permit number 279652 and CITES import permit number 563301. Details pertaining to the locations of animals used in this study are not disclosed due to the confidentiality of black rhino data which is a policy of the IUCN SSC African Rhino Specialist Group.
Study populations
The EAZA Endangered Species Programme (EEP) for the eastern black rhino is maintained in European zoos and managed using a detailed studbook. The studbook contains comprehensive records dating back to the early 1960s, while some lineages can be traced back to 1948. The population currently numbers around ~ 90 individuals (Pilgrim, pers. comm.) and breeding recommendations are made using a mean-kinship strategy (Lacy et al. 2011).
The second ex situ population (SAx) is extralimital and now held on a private reserve in South Africa. The founding animals were captured in Kiboko, Kenya, in 1961, and translocated to Addo National Park. This population was intended as an insurance population at a time when Kenya’s black rhino were threatened by steep decline (Emslie & Brooks 1999; Muya et al. 2011; Okita-Ouma et al. 2007). However, three males of a divergent ESU, the southern black rhino (D. b. bicornis), were transferred into the ex situ population while in Addo National Park in 1977 (Hall-Martin 1984). As a result, the population contains an unknown proportion of southern admixture (Kretzschmar, pers. comm.). The original intention of this insurance population was to return SAx descendants to Tsavo National Park to supplement the small in situ population which had declined to only 15 animals in 1991 (Hall-Martin 1991). However, admixed animals may not be favourable for translocation into native populations in East Africa.
Sampling and genotyping
Samples were collected from zoo animals, between 2017 and 2021, using non-invasive nasal swabs (N = 75), or whole blood (N = 10) where this could be obtained opportunistically. One muscle tissue sample from a deceased animal, preserved in 2ml absolute ethanol at -20°C, was also included. Nasal swabs were preserved using silica gel at room temperature and blood samples collected in EDTA tubes were frozen at -20°C. From the SAx population we extracted DNA from pinna offcuts preserved in sodium chloride. These were obtained during routine ear-notching operations performed to facilitate individual animal identification on the reserve. Samples used in this study dated up to 2018, but samples from historic assignments and some adults were not available. We sampled approximately 80% of the SAx and 85% of the EEP population.
DNA was extracted using a QIAamp DNA mini kit (QIAGEN GMBH, Hilden, Germany). Mitochondrial DNA was amplified using D-loop primers mt15996L and mt16502H following Moodley et al., (2017). Individuals were Sanger sequenced in the forward and reverse direction (Eurofins Genomics Europe) and consensus sequences generated using Geneious software (Geneious Prime 2023.0.1, https://www.geneious.com). These sequences were aligned with the corresponding homologous sequenced region from the Moodley et al., (2017) metapopulation database and a haplotype network was constructed using PopArt (Version 1.7, Leigh & Bryant 2015).
We genotyped each sample with 15 polymorphic autosomal microsatellite loci isolated from the black rhino (Diceros bicornis) and white rhino (Ceratotherium simum). This included 10 of the 11 loci applied in Moodley et al., (2017). We included additional markers DB49, DB66, and BlRh1B (Brown & Houlden 1999), and BlRh1C and DB52 (Harper et al. 2013). We followed the protocol outlined in Moodley et al., (2017). The additional primer sets were amplified as multiplexes using annealing temperatures of 57 °C (DB49, DB66, BlRh1B) and 52 °C (BlRh1C and DB52) (see SI Table 1). Samples were analysed on a 48-well capillary ABI3730 genetic analyser (Applied Biosystems) with Genescan ROX500 size standards (Applied Biosystems). Allele sizes were determined using using the Geneious software microsatellite plugin (Geneious Prime 2023.0.1, https://www.geneious.com). We tested the markers for linkage disequilibrium using the R package pegas (Paradis 2010). All R analyses were conducted using R (version 4.3.1, R Development Core Team, 2011) and RStudio (version 2023.09.1.494, Posit team 2023).
Using the 15 markers we calculated mean allelic richness (AR), and observed and expected heterozygosity (HO and HE, respectively) for each population using R packages PopGenReport and Poppr (Adamack and Gruber 2014; Kamvar et al. 2014; Adamack & Gruber 2014; Kamvar et al. 2014) Population level differentiation, FST, and the inbreeding coefficient, FIS, were calculated using a Weir and Cockerham estimator in R package hierfstat (Weir & Goudet 2017). We estimated the effective population sizes for the EEP and SAx populations using the linkage disequilibrium method, under random mating and with a critical minor allele frequency of 0.05, implemented in NeEstimator v2 (Do et al. 2014).
Pedigree reconstruction
Pedigree reconstruction for the EEP and SAx was carried out on the microsatellite dataset using FRANz (version 2.0.0, Riester et al. 2009). Pedigree reconstruction was implemented for each population separately by adding observed mothers in a stepwise fashion to the FRANz input file. For the EEP, we ran FRANz without the full-sib heuristic, with a female reproductive span of 5 to 30 years of age, a male reproductive span of 5 to 40, and ran the simulation for 1,000,000 iterations. We first ran FRANz without any known relationships included and retained any consensus assignments to known mothers in the second model, to which we then also added mother–offspring pairs with the rarest mtDNA genotypes. After each run, we revised the input file of known relationships to remove any parents with a posterior probability of < 0.8 and added further known mothers based on the frequency of the mtDNA genotype in the population. We averaged the posterior probability for each parent over 20 repeat runs once all mothers were added and removed any assignment with a probability < 0.8. We compared the resulting pedigree to studbook records and counted the number of erroneous or missing assignments to benchmark this method. Marker DB49 showed the highest number of mismatches with the studbook, but removing this marker did not improve the accuracy of pedigree reconstruction in comparison with the studbook data.
For the SAx population we repeated the above mentioned process. A mother–offspring studbook has been curated from observational data collected during ear-notching operations. We first used mitochondrial (mtDNA) genotypes to remove any erroneous mother–offspring relationships. Where a different mother to the observed mother was assigned, the microsatellite genotypes of the candidate parents were compared with the offspring and any assigned father to check for correct assignment. Finally, we added additional parents for unsampled individuals to increase network connectivity. These were based on known relationships from observational data (mother-calf relationships, (Hall-Martin 1986), or previous genetic relationships (Harper pers. comm., unpublished data, SI Table 2). We ran the final configuration 20 times to generate consensus probabilities for all parent offspring pairs and eliminate spurious assignments. The resulting pedigree network was visualised using Helium (Shaw et al. 2014).
Quantifying admixture
In order to quantify the proportion of southern ancestry across the pedigrees we joined the reconstructed SAx pedigree produced by FRANz, with historic relationships assigned from a combination of historic records (Hall-Martin 1986) and molecular data (Harper pers. comm.). Based on the early assignments among population founders we used the R package OptiSelect (Wellmann et al. 2012) to quantify the contribution of each male founder to both the zoo and SAx pedigrees.
Evaluating reproductive skew
Here we consider that reproductive skew is a result of the intrinsic (individual fitness) and the extrinsic (management strategy) components. To evaluate variation in reproductive skew and genetic drift between the two populations, we used the breeding histories from the studbook and the reconstructed pedigree. For the EEP we restricted our analyses to the 32-year time period between 1990 and 2022 to reflect the current management strategy. We excluded any offspring that were stillborn or did not reach one year of age. We compiled a list of candidate parents including all animals that were alive in the EEP between 1990 and 2022 and were > 5 years of age (Brodie et al. 2011). Animals were excluded as candidate parents on death or on transfer out of the zoo breeding population. For each candidate parent we recorded the years they were assigned as sire or dam in the list of offspring born between 1990 and 2022.
For the SAx population we restricted our analyses to the period between 2003 and 2018 as the last males were translocated into the population in 2001 and black rhino have a gestation period of a minimum of 15 to 18 months (Le Roex & Ferreira 2020; Okita-Ouma et al. 2021). To compare measures of skew to the zoo population we used a minimum age of five years for candidate parents, even though free-ranging males tend not to reproduce successfully until they are competitive at nine or ten years of age (Law & Linklater 2014). In the SAx population some males are located on an adjoining reserve to reduce conflict. Males were occasionally transferred between these two populations or at times held in an enclosure with females. We included all animals aged > 5 as candidate parents to reflect the management strategy. We used the reconstructed pedigree to list the births assigned to each candidate parent. For each population, we calculated the multinomial index of reproductive skew, using the R package SkewCalc (Ross et al. 2020), which accounts for a non-linear relationship between age and reproductive success. If < 5% of the posterior distribution of M falls within the Region of Practical Equivalence (ROPE) of − 0.1 to 0.1, the null hypothesis of no skew can be rejected.
To evaluate how well each population has preserved founder genetic diversity we quantified the contribution of each founder to the current population (2022) for males and females from zoos, and the contributions of male founders to the current and total SAx population. We were unable to make accurate estimations for female founders from the SAx population as early relationships in the pedigree were unassigned for animals that were not sampled in this study.
Results
Mitochondrial diversity
A total of 177 (SAx = 103, EEP = 74) mitochondrial d-loop consensus sequences of length 456 bp were obtained, featuring 28 polymorphic sites (Genbank OR709675—OR709685). We identified 10 haplotypes, of which 3 were shared between the EEP and SAx, one was unique to SAx, and 6 haplotypes were unique to the EEP. Two of the EEP haplotypes fall within the southern ESU (Fig. 1, SN; Moodley et al. 2017), the first of which is from a male ‘Kalusho’ from the discontinued D. b. minor EEP, and the second from a D. b. michaeli EEP male ‘Sammy’ indicating southern maternal ancestry.
Median-joining haplotype network of mitochondrial d-loop sequences. Haplotypes from the two ex situ populations are presented in the context of the data and evolutionary groups from the Moodley et al. (2017) meta-population study. Broadly, the groups Eastern (EA) and Central (CE) represent an eastern cluster (blue shading), and groups South-Eastern (SE), South-Northern (SN) and South-Western (SW) represent a southern cluster (gold shading). All maternal lineages bar two EEP animals have an eastern origin. Of these two southern haplotypes in the zoo population, one is an individual from the discontinued D. b minor breeding programme, and one is an admixed male with maternal southern ancestry
All of the four SAx haplotypes had an eastern origin, confirmed by comparisons with the Moodley et al., (2017) historic mtDNA dataset (Fig. 1). This low number reflects the small number of female founders. Based on their relationships in the pedigree we were able to infer that the original female founders ‘Ida’ and ‘Brunni’ share an eastern mtDNA haplotype. This is likely due to the close proximity of their capture sites in Kiboko, Kenya and indicates a common ancestor. A second mitochondrial haplotype was associated with the lineage of female ‘Kate’. Of the final two haplotypes found in the SAx population, one was found only in a male founder from Tanzania, ‘Richard’ and one originated from the EEP population through female ‘Tana’, one of two captive-born females translocated from Port Lympne Safari Park in the UK to the SAx population in 2004 (King & Beer 2018).
Microsatellite diversity
Across both populations, 195 individuals (EEP N = 85, SA-x N = 110) amplified reliably at a minimum of 12 of the 15 polymorphic microsatellite loci. The mean number of alleles per locus was 7.27 ± 2.49 ranging from 4 alleles (DB23, DB14, SW35) to 11 alleles (Br4, DB66).
Both populations had similar expected and observed heterozygosity (EEP: HE = 0.665, HO = 0.706, SAx: HE = 0.673, HO = 0.699) (Table 1). Private alleles were higher in the EEP (28) than in SAx (6) consistent with the larger number of founders. Population FIS was not significantly different from zero in both ex situ populations (EEP -0.062, SAx—0.039), indicating no excess of inbreeding. Ne was estimated at 23.3 and 12.8 for the EEP and SAx populations respectively using the linkage disequilibrium method (Neestimator v2, Do et al. 2014). The two populations showed weak differentiation with an FST value of 0.029.
Validating pedigree reconstruction using the EEP studbook
Based on 48 known maternal relationships, 30 known paternal relationships were correctly generated by FRANz with reference to the studbook data. 78 relationships were correctly assigned as missing, as the parent was not sampled in our dataset. Eight paternal relationships were unassigned despite the father being present in the dataset with a posterior probability of < 0.8. Four parent–offspring relationships were incorrectly assigned by FRANz with two being assigned to full-siblings and two to half-siblings.
Reconstructing the pedigree of the SAx population
The proposed pedigree includes a total of 168 relationships within the population. 77 mother–offspring relationships were assigned based on our molecular data, of which 69 confirmed observational assignments. FRANz highlighted eight false assignments in the observational pedigree based on > 2 mismatched markers between mother and offspring genotypes. In two of these cases the correct mother was able to be assigned with molecular data, whereas for the other mismatches the mothers were presumably not sampled. We were able to assign paternity for 68 animals based on microsatellite data. Five impossible assignments resulting from an unsampled father were removed due to either spatial considerations (animals held on different parts of the reserve), or the very young age of the candidate father (6 years old). Additional relationships in the pedigree were added from observed assignments or previous assignments from molecular data which were not genotyped in this study (Kretzschmar, pers comm.). None of these additional assignments caused the pedigree to ‘break’, but these relationships should be considered candidates rather than confirmed (SI Table 1). For a visualization of the pedigree, see Fig. 2.
Complex pedigree of the SAx population. Females are represented by light blue, and males by light green circles. Maternal relationships are indicated by red lines and paternal ones by blue lines. Relationships above the dotted line are derived from observational records from the SAx population while it was at Addo Elephant National Park (Hall-Martin 1986) and some assignments are based on molecular data generated by Harper (unpublished data). Individuals below the black line are assignments from this study. Where samples weren’t available for genotyping, for some individuals, paternity was inferred by excluding sampled candidate fathers
Free-ranging female black rhino may produce their first offspring aged five or six (Okita-Ouma et al. 2021), the earliest age of reproduction documented in the SAx population was six years old (N = 3). Most females will have produced an offspring by age nine (Harvey Sky et al. 2022). Of the 25 sampled candidate mothers aged over six at the birth of the youngest animal, 18 accounted for the 74 births for which maternity could be assigned. Four animals in the candidate parent group had not reproduced by the age of 9. The average inter-calving interval (ICI) for females ranged from 2 to 3.33 years, and four females had an average inter-calving interval < 2.5 years indicating a highly productive population.
Male black rhino do not become reproductively competitive until the age of nine or ten (Adcock 1994). For genotyped males aged > 10 at the approximate conception time of the most recent cohort, this span included 67 births spread over 22 of the 26 candidate fathers. Unfortunately, samples were not available for all the population, therefore genotypes for several animals are missing. One ungenotyped male ‘Bwana’ is suggested to have contributed significantly to the population based on observations. As a result, one of his offspring was assigned a high proportion of the paternal relationships by FRANz. This was subsequently corrected based on the offspring’s young age (< 6 years old) and Bwana was listed as the likely parent.
Reproductive skew
In a model of the multinomial index of reproductive skew, M, accounting for animal age, there was no skew present among females in either population with the posterior probability distribution overlapping zero (Fig. 3, SAx: median 0.06, 95% CI -0.42 to 0.56, 37.74% in ROPE; EEP: median 0.10, 95% CI -0.25 to 0.54, 39.95% in ROPE). Zoo males showed some skew with a median of 0.59, 95% CI 0.09 to 1.38, 0.42% in ROPE. Skew was, however, high among SAx males (median 1.31, 95% CI 0.21 to 3.10, 0% in ROPE) and significantly higher than among zoo males (median of difference 0.72, 95% CI 0.12 to 1.72, 0% in ROPE).
Multinomial index of reproductive skew (M) for males and females, accounting for age. No skew was present for females in either populations as distributions were not different from zero. Skew is found among males in both populations, but is significantly higher for the free-ranging SAx population than for the European zoo population which is managed by mean-kinship breeding (median of difference 0.72, 95% CI 0.12 to 1.72)
Founder contributions
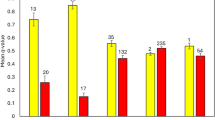
Under an ideal scenario, where drift is inconsequential, each founder lineage would have equal representation in the population. For the zoo population, founder contributions among females varied from 17.3% (Jimmi, studbook number 175, Tsavo National Park, Kenya) to 0.6% (June, studbook number 17, unknown origin; Jarca, studbook number 178, Tsavo, Tanzania, Fig. 4A). For male founders of the European zoo population, contributions also varied, ranging from 13.1% (Gareth Edwards, studbook number 534, from SAx (Addo) South Africa) to 0.7% (Conni, studbook number 36, recorded as from ‘Kenya Game Ward’, Fig. 4A). Although Gareth Edwards originated in the SAx population, based on his assigned relationships he is predicted to be unadmixed. Conversely, male Madiba who also originates in SAx is estimated to have 25% southern ancestry by the paternal relationship assigned from the pedigree reconstruction. Likewise, an examination of records compiled by Rookmaaker et al. (1998), revealed that male ‘Sammy’, imported from Japan has 50% southern ancestry with his maternal grandparents both imported from Namibia (Rookmaaker et al. 1998). Consequently, while the genetic makeup of the European zoo population is predominantly eastern black rhino, admixture is also present (3.5% of the current population).
A Contributions (%) of each founder to the current European zoo population based on the studbook. For founders with lower contributions, these lineages are likely to be lost from the population. B contributions (%) of each male founder to the current SAx population. The southern male founder has contributed the greatest population of ancestry to the SAx population
In the SAx population, founder contributions among males show strong asymmetry (Fig. 4B) with one of the original founders ‘Darkie’ contributing only 9.6% of total male ancestry (Table 2). The other original male founder from Kenya, ‘J.A.’ has contributed a higher proportion of ancestry (22.76%) while more recent founders ‘Bwana’ translocated from Port Lympne Safari Park in the UK in 1995, and ‘Richard’ from Tanzania contributed intermediate proportions of ancestry (12.57% and 15.72%, respectively). However, the highest proportion of ancestry is assigned to the southern black rhino male (SBR, 39.35%, Table 2).
Actual values may deviate slightly due to recombination and the random segregation of gametes. The southern black rhino male has contributed the greatest proportion of male ancestry to the SAx population. Subspecies assignments are inferred based on capture location.
Discussion
Following a decline of over 96% in the wild (Harley et al. 2005), the two ex situ populations examined here represent an important and substantial proportion of the global black rhino meta-population. In this study we have reconstructed the pedigree of the freely mixing SAx population with a high degree of certainty that enabled a reliable assessment of reproductive skew and the contribution of southern ancestry. This has enabled a real-world comparison of two contrasting management strategies commonly employed for endangered species. Elucidating the genetic effects of population management in black rhino, in contrast to animals with a shorter generation time (e.g. the Tasmanian devil, Sarcophilus harrisii, Farquharson et al. 2021; Wright et al. 2020), will have wider implications for how we manage meta-populations of other Critically Endangered species.
A detailed understanding of diversity, vulnerability to drift, and individual proportions of admixture in these two key populations offers exceptional scope for detailed management strategies that will maximise their conservation value and begin to bridge the gap between ex situ and in situ conservation.
The SAx population has reasonable levels of diversity in spite of its small number of founders. However, the European zoo population retains a higher diversity of microsatellite alleles and mtDNA haplotypes. Indeed, the European zoos contain high genetic diversity based on microsatellite genotypes, comparable to the diversity observed in Kenya (HO = 0.73 ± 0.03, Muya et al. 2011), and substantially higher than estimates for the southern black rhino (HO = 0.436, Harley et al. 2005) and the south-western black rhino (HO = 0.523, Harley et al. 2005).
Maintaining this diversity over time requires the effects of genetic drift to be minimised so that rare variants are more likely to persist over generations. In this study we show that the mean-kinship breeding strategy in the zoo effectively reduces reproductive skew in comparison to a freely mixing population with low intervention. Both the reduction in reproductive skew and the larger population of founders are reflected in the larger effective population size of the European zoos. Conversely, the SAx population has a small effective population size which, with sustained high reproductive skew, means this population will be subjected to stronger effects of genetic drift and will likely lose diversity more quickly. Increasing the effective population size must be a priority given the precarious future predicted by our findings if this population were to remain genetically isolated.
Acting to counter drift by maximizing the effective population size is most important when the population is small. Selection becomes more effective and drift less extreme as the population becomes larger. In comparison to the effective population size of large and connected historic populations, both of these ex situ populations would be considered small, but there is also an argument to be made to maintain the semi-natural breeding system and associated level of reproductive skew in the context of the SAx population. Implementing a mean-kinship breeding strategy circumvents reproductive selection, which is maintained in the SAx population, as to an extent are many ‘natural’ ecological processes. Altered selection can result in inefficient purifying selection against mutations that may be deleterious in situ, and an increase in the frequency of these mutations may compromise fitness (Wilder et al. 2020) and the success of reintroductions.
Adaptation to captivity, is a major concern for ex situ populations (Frankham 2008), and a particular concern for populations in zoos. Equalizing contributions to the subsequent generation by mean-kinship breeding, rather than selecting for fecundity, should function to slow the rate of adaptation to captivity (Williams & Hoffman 2009), but purifying selection may still be relaxed. Although the mean-kinship breeding programme is effective at reducing the current degree of reproductive skew in the zoo population, founder contributions in this population are highly unequal. Moreover, Edwards et al. (2015a, b) recorded a potential founder population size of 135 wild-caught individuals, of which only 41 have contributed to the extant zoo population, representing a loss of 69% of potential lineages. Nonetheless, we found no evidence of reproductive skew for females in subsequent generations. It is possible that this later lack of skew reflects improved husbandry, or alternatively early variation in reproductive success may have selected for lineages that do well in captivity. Interestingly, skew was still present among males in zoos. This may reflect the fact that males are often favoured over females for translocations intended to inject new diversity into a breeding programme. Some males will therefore reflect more unique lineages and will be favoured by the mean-kinship breeding strategy due to their low relatedness to the population. Unfortunately, two of these imported males ‘Sammy’ and ‘Madiba’ are admixed and this has resulted in the spread of a low proportion of southern ancestry in the European zoo population, although currently this is confined to certain lineages.
Indeed, the diverse founder composition of the zoo population, primarily originating in East Africa, has likely resulted in the interbreeding of several eastern evolutionary units (Sánchez-Barreiro et al. 2023). As a rule, the translocation of admixed animals from ex situ populations into existing native populations in situ is a high-risk strategy and is actively discouraged (Bertola et al. 2022). However, this may not preclude translocations of zoo animals from mixed eastern ESUs into the native range, given that a similar mixing of animals has likely also taken place in Kenya (Muya et al. 2011). The zoo population may therefore serve as a useful reservoir of diversity for future translocations, even where native eastern populations persist, but it is key that any translocations be genomically informed to prevent any further erosion of rare native diversity.
The conservation value of eastern black rhino with southern ancestry remains an open question. Admixture between subspecies can result in increased fitness of hybrid offspring through heterosis due to the masking of deleterious alleles (Onley et al. 2022; Wei & Zhang 2018). However, outbreeding depression may result from the breakdown of co-adapted gene-complexes (Edmands & Timmerman 2003; Frankham et al. 2011). This gamble is not merited by the current status of the black rhino, particularly considering the deep 641Ka divergence between eastern and southern clades (Moodley et al. 2020). This divergence far exceeds the recommendations regarding admixture which are intended to minimize the risk of outbreeding depression (Frankham et al. 2011).
In the zoo population, southern admixture is recent and limited, therefore pairing decisions could be made to prevent further spread of southern alleles. However, the situation in the SAx population is more complex. Admixture from the subspecies D. b. bicornis is extensive in this population, but not reflected among mitochondrial haplotypes due to its paternal origin. Indeed, the southern male, has contributed a greater proportion of ancestry than any other male founder. This southern male, added to the population on Addo, in 1977, to replace the dominant eastern bull after his death (Hall-Martin 1984), was the only breeding male over several years at a time when the population was small (ca. 15 individuals). This proliferation of southern ancestry is an accident of timing and historic management rather than sexual selection but has led to the extensive introgression of southern ancestry throughout the SAx population. Management strategies including the translocation of eastern males into the population are currently being implemented to reduce the proportion of admixture and increase Ne, but the translocation of admixed animals continues to pose a dilemma.
The original intent of the SAx population was to return animals to where the eastern founders were captured, but the growth of the population in the Tsavo region of Kenya (Chyulu hills, Tsavo East, Tsavo West, Ngulia and the Intensive Protection Zone (IPZ)) to around 363 black rhino (Kenya Wildlife Service 2022), means translocations of admixed SAx animals would be better prioritized elsewhere. Contractually the SAx animals may not be translocated elsewhere in South Africa, being regarded as an alien subspecies (Emslie 2020). Translocations to Tanzania have already been undertaken (Kretzschmar, pers. comm.), however a better strategy may be to introduce these animals to regions where admixture may have historically occurred at the boundary of the D. b. michaeli and D. b. bicornis range in Malawi, Zambia and Southern DRC. Alternatively, these animals would be a potential source for the founding of new populations where black rhino are regionally extinct, as is the case for recent translocations to Rwanda and Chad (Bantlin, pers. comm.)
This study contributes to the relatively small number of detailed studies on genetic management (e.g. Tasmanian devil (Sarcophilus harrisii), Farquharson et al. 2021; Wright et al. 2020; Kākāpo (Strigops habroptilus), Guhlin et al. 2023). Our findings therefore have wider implications for how we manage meta-populations of other Critically Endangered species. Firstly, generating reliable data on genetic diversity and reproductive skew under different management strategies can inform decisions on the optimal means to ensure long-term maintenance of genetic diversity in small human-modified populations. Secondly, as for the black rhino, admixture is a frequent outcome of unknown founder diversity in many ex situ populations (Hvilsom et al. 2013; Węcek et al. 2016). This study demonstrates the importance of tracing and identifying ancestry to inform appropriate translocations, integrate ex situ populations into meta-population level strategy and realise their conservation potential. By incorporating genetic information into translocation strategy, risk can be minimized, translocation success improved and the long-term benefits from these costly exercises maximized.
Data availability
Unique mitochondrial haplotype data are deposited to NCBI Nucleotide Database (Genbank accession numbers OR709675—OR709685). Individual microsatellite genotype data and mitochondrial haplotype data are available on DataDryad (https://doi.org/10.5061/dryad.69p8cz97p). In accordance with AfRSG guidelines, detailed meta-data for black rhino are not shared.
References
Adamack AT, Gruber B (2014) PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol Evol 5(4):384–387. https://doi.org/10.1111/2041-210X.12158
Adams JR, Vucetich LM, Hedrick PW, Peterson RO, Vucetich JA (2011) Genomic sweep and potential genetic rescue during limiting environmental conditions in an isolated wolf population. Proc R Soc B 278(1723):3336–3344. https://doi.org/10.1098/rspb.2011.0261
Adcock, K. (1994). The relevance of “territorial” behaviour in black rhino to their population management. Proceedings of a Symposium on Rhinos as Game Ranch Animals. Onderstepoort, South Africa.
Bergner LM, Jamieson IG, Robertson BC (2014) Combining genetic data to identify relatedness among founders in a genetically depauperate parrot, the Kakapo (Strigops habroptilus). Conserv Genet 15(5):1013–1020. https://doi.org/10.1007/s10592-014-0595-y
Bertola LD, Miller SM, Williams VL, Naude VN, Coals P, Dures SG, Henschel P, Chege M, Sogbohossou EA, Ndiaye A, Kiki M, Gaylard A, Ikanda DK, Becker MS, Lindsey P (2022) Genetic guidelines for translocations: Maintaining intraspecific diversity in the lion (Panthera leo). Evol Appl 15(1):22–39. https://doi.org/10.1111/eva.13318
Bogoni JA, Peres CA, Ferraz KMPMB (2020) Extent, intensity and drivers of mammal defaunation: a continental-scale analysis across the Neotropics. Sci Rep 10(1):14750. https://doi.org/10.1038/s41598-020-72010-w
Britnell JA, Zhu Y, Kerley GIH, Shultz S (2023) Ecological marginalization is widespread and increases extinction risk in mammals. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.2205315120
Brodie JF, Muntifering J, Hearn M, Loutit B, Loutit R, Brell B, Uri-Khob S, Leader-Williams N, du Preez P (2011) Population recovery of black rhinoceros in north-west Namibia following poaching. Anim Conserv 14(4):354–362. https://doi.org/10.1111/j.1469-1795.2010.00434.x
Brown SM, Houlden BA (1999) Isolation and characterization of microsatellite markers in the black rhinoceros (Diceros bicornis). Mol Ecol 8(9):1559–1561. https://doi.org/10.1046/j.1365-294X.1999.07246.x
Cain B, Wandera AB, Shawcross SG, Edwin Harris W, Stevens-Wood B, Kemp SJ, Okita-Ouma B, Watts PC (2014) Sex-biased inbreeding effects on reproductive success and home range size of the critically endangered black rhinoceros. Conserv Biol 28(2):594–603. https://doi.org/10.1111/cobi.12175
Cunningham J, Harley EH, O’Ryan C (1999) Isolation and characterization of microsatellite loci in black rhinoceros (Diceros bicornis). Electrophoresis 20(8):1778–1780. https://doi.org/10.1002/(SICI)1522-2683(19990101)20:8%3c1778::AID-ELPS1778%3e3.0.CO;2-W
DeWoody JA, Harder AM, Mathur S, Willoughby JR (2021) The long-standing significance of genetic diversity in conservation. Mol Ecol 30(17):4147–4154. https://doi.org/10.1111/mec.16051
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 14(1):209–214. https://doi.org/10.1111/1755-0998.12157
Edmands S, Timmerman CC (2003) Modeling factors affecting the severity of outbreeding depression. Conserv Biol 17(3):883–892. https://doi.org/10.1046/j.1523-1739.2003.02026.x
Edwards KL, Shultz S, Pilgrim M, Walker SL (2015a) Irregular ovarian activity, body condition and behavioural differences are associated with reproductive success in female eastern black rhinoceros (Diceros bicornis michaeli). Gen Comp Endocrinol 214:186–194. https://doi.org/10.1016/j.ygcen.2014.07.026
Edwards KL, Walker SL, Dunham AE, Pilgrim M, Okita-Ouma B, Shultz S (2015b) Low birth rates and reproductive skew limit the viability of Europe’s captive eastern black rhinoceros. Diceros Bicornis Michaeli Biodiv Conserv 24(11):2831–2852. https://doi.org/10.1007/s10531-015-0976-7
Emslie, R., & Brooks, M. (1999). African rhino: status survey and conservation action plan.
Emslie, R. (2020). Diceros bicornis. The IUCN Red List of Threatened Species 2020. https://doi.org/10.2305/IUCN.UK.2020-1.RLTS.T6557A152728945.en. Available at: https://www.iucnredlist.org/species/6557/152728945
Farquharson KA, McLennan EA, Wayne A, Smith M, Peel E, Belov K, Hogg CJ (2021) Metapopulation management of a critically endangered marsupial in the age of genomics. Global Ecol Conserv 31:e01869. https://doi.org/10.1016/j.gecco.2021.e01869
Farré M, Johnstone C, Hopper J, Kitchener AC, Roos C, King T (2022) Novel mtDNA haplotypes represented in the European captive population of the Endangered François’ langur (Trachypithecus francoisi). Int J Primatol 43(3):533–537. https://doi.org/10.1007/s10764-022-00295-x
Florescu A, Davila JA, Scott C, Fernando P, Kellner K, Morales JC, Melnick D, Boag PT, Groot VCD, P. (2003) Polymorphic microsatellites in white rhinoceros. Mol Ecol Notes 3(3):344–345. https://doi.org/10.1046/j.1471-8286.2003.00440.x
Frankham R (2008) Genetic adaptation to captivity in species conservation programs. Mol Ecol 17(1):325–333. https://doi.org/10.1111/j.1365-294X.2007.03399.x
Frankham R, Ballou JD, Eldridge MDB, Lacy RC, Ralls K, Dudash MR, Fenster CB (2011) Predicting the probability of outbreeding depression. Conserv Biol 25(3):465–475. https://doi.org/10.1111/j.1523-1739.2011.01662.x
Galla SJ, Brown L, Couch-Lewis (Ngāi Tahu: Te Hapū o Ngāti Wheke, N. W. Y., Cubrinovska, I., Eason, D., Gooley, R. M., Hamilton, J. A., Heath, J. A., Hauser, S. S., Latch, E. K., Matocq, M. D., Richardson, A., Wold, J. R., Hogg, C. J., Santure, A. W., & Steeves, T. E. (2022) The relevance of pedigrees in the conservation genomics era. Mol Ecol 31(1):41–54. https://doi.org/10.1111/mec.16192
García-Dorado A, Caballero A (2021) Neutral genetic diversity as a useful tool for conservation biology. Conserv Genet 22(4):541–545. https://doi.org/10.1007/s10592-021-01384-9
Garnier JN, Bruford MW, Goossens B (2001) Mating system and reproductive skew in the black rhinoceros. Mol Ecol 10(8):2031–2041. https://doi.org/10.1046/j.0962-1083.2001.01338.x
Gibson I, Welsh AB, Welsh SA, Cincotta DA (2019) Genetic swamping and possible species collapse: tracking introgression between the native Candy Darter and introduced Variegate Darter. Conserv Genet 20(2):287–298. https://doi.org/10.1007/s10592-018-1131-2
Gooley RM, Hogg CJ, Belov K, Grueber CE (2018) The effects of group versus intensive housing on the retention of genetic diversity in insurance populations. BMC Zoology 3(1):2. https://doi.org/10.1186/s40850-017-0026-x
Guhlin J, Le Lec MF, Wold J, Koot E, Winter D, Biggs PJ, Galla SJ, Urban L, Foster Y, Cox MP, Digby A, Uddstrom LR, Eason D, Vercoe D, Davis T, Andrew K, Argilla L, Arnold K, Bohan J, Dearden PK (2023) Species-wide genomics of kākāpō provides tools to accelerate recovery. Nat Ecol Evol 7(10):1693–1705
Hall-Martin A (1984) Kenya’s black rhino in Addo, S Africa. Newsletter African Elephant Rhino Group 3:11
Hall-Martin A (1991) Adding to Addo. Rhino and Elephant Foundation Journal 5:18–20
Hall-Martin, A. J. (1986). Recruitment in a Small Black Rhino Population. Pachyderm, No. 7, 6–8. figs. 1–3, tables 1–3.
Harley EH, Baumgarten I, Cunningham J, O’Ryan C (2005) Genetic variation and population structure in remnant populations of black rhinoceros, Diceros bicornis, in Africa. Mol Ecol 14(10):2981–2990. https://doi.org/10.1111/j.1365-294X.2005.02660.x
Harper CK, Vermeulen GJ, Clarke AB, de Wet JI, Guthrie AJ (2013) Extraction of nuclear DNA from rhinoceros horn and characterization of DNA profiling systems for white (Ceratotherium simum) and black (Diceros bicornis) rhinoceros. Forensic Sci Int Genet 7(4):428–433. https://doi.org/10.1016/j.fsigen.2013.04.003
Harvey Sky N, Jackson J, Chege G, Gaymer J, Kimiti D, Mutisya S, Nakito S, Shultz S (2022) Female reproductive skew exacerbates the extinction risk from poaching in the eastern black rhino. Proc Royal Soc B. https://doi.org/10.1098/rspb.2022.0075
Hogg CJ, Wright B, Morris KM, Lee AV, Ivy JA, Grueber CE, Belov K (2019) Founder relationships and conservation management: empirical kinships reveal the effect on breeding programmes when founders are assumed to be unrelated. Anim Conserv 22(4):348–361. https://doi.org/10.1111/acv.12463
Hohenlohe PA, Funk WC, Rajora OP (2021) Population genomics for wildlife conservation and management. Mol Ecol 30(1):62–82. https://doi.org/10.1111/mec.15720
Howard JG, Lynch C, Santymire RM, Marinari PE, Wildt DE (2016) Recovery of gene diversity using long-term cryopreserved spermatozoa and artificial insemination in the endangered black-footed ferret. Anim Conserv 19(2):102–111. https://doi.org/10.1111/acv.12229
Hvilsom C, Frandsen P, Børsting C, Carlsen F, Sallé B, Simonsen BT, Siegismund HR (2013) Understanding geographic origins and history of admixture among chimpanzees in European zoos, with implications for future breeding programmes. Heredity 110(6):586–593. https://doi.org/10.1038/hdy.2013.9
Ivy JA, Lacy RC (2012) A comparison of strategies for selecting breeding pairs to maximize genetic diversity retention in managed populations. J Hered 103(2):186–196. https://doi.org/10.1093/jhered/esr129
Kamvar ZN, Tabima JF, Grünwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. https://doi.org/10.7717/peerj.281
Kenya Wildlife Service. (2022). Recovery and Action Plan for the Black Rhino in Kenya (2022–2026).
King, T., & Beer, P. (2018). Rhinos, elephants and The Aspinall Foundation: over 30 years of captive-breeding, reintroduction and conservation. Pachyderm, 59, 127–131. https://pachydermjournal.org/index.php/pachyderm/article/view/96
Lacy R, Ballou J, Pollak J (2011) PMx: software package for demographic and genetic analysis and management of pedigreed populations. Methods Ecol Evol 3(2):433–437. https://doi.org/10.1111/j.2041-210X.2011.00148.x
Law PR, Linklater WL (2014) Black rhinoceros demography should be stage, not age, based. Afr J Ecol 52(4):571–573. https://doi.org/10.1111/aje.12148
Law PR, Fike B, Lent PC (2013) Mortality and female fecundity in an expanding black rhinoceros (Diceros bicornis minor) population. Eur J Wildl Res 59(4):477–485. https://doi.org/10.1007/s10344-013-0694-y
Le Roex N, Ferreira SM (2020) Age structure changes indicate direct and indirect population impacts in illegally harvested black rhino. PLoS ONE 15(7):e0236790. https://doi.org/10.1371/journal.pone.0236790
Leigh JW, Bryant D (2015) popart : full-feature software for haplotype network construction. Methods Ecol Evol 6(9):1110–1116. https://doi.org/10.1111/2041-210X.12410
Leus K, Traylor-Holzer K, Lacy RC (2011) Genetic and demographic population management in zoos and aquariums: Recent developments, future challenges and opportunities for scientific research. International Zoo Yearbook 45(1):213–225. https://doi.org/10.1111/j.1748-1090.2011.00138.x
Miller-Butterworth CM, Vacco K, Russell AL, Gaspard JC (2021) Genetic Diversity and Relatedness among Captive African Painted Dogs in North America. Genes 12(10):1463. https://doi.org/10.3390/genes12101463
Modesto P, Biolatti C, Favaro L, Colussi S, Peletto S, Piga S, Riina MV, Pessani D, Trincas E, Isaja V, Acutis PL (2018) Molecular Genetics Unveiled Unknown Family Relationships and Hybrids in an Ex-Situ Colony of African Penguins (Spheniscus demersus). J Hered 109(6):653–662. https://doi.org/10.1093/jhered/esy032
Moodley Y, Russo IRM, Dalton DL, Kotzé A, Muya S, Haubensak P, Bálint B, Munimanda GK, Deimel C, Setzer A, Dicks K, Herzig-Straschil B, Kalthoff DC, Siegismund HR, Robovský J, O’Donoghue P, Bruford MW (2017) Extinctions, genetic erosion and conservation options for the black rhinoceros (Diceros bicornis). Sci Rep 7(February):1–16. https://doi.org/10.1038/srep41417
Moodley Y, Westbury MV, Russo I-RM, Gopalakrishnan S, Rakotoarivelo A, Olsen R-A, Prost S, Tunstall T, Ryder OA, Dalén L, Bruford MW (2020) Interspecific gene flow and the evolution of specialization in black and white rhinoceros. Mol Biol Evol 37(11):3105–3117. https://doi.org/10.1093/molbev/msaa148
Moran BM, Thomas SM, Judson JM, Navarro A, Davis H, Sidak-Loftis L, Korody M, Mace M, Ralls K, Callicrate T, Ryder OA, Chemnick LG, Steiner CC (2021) Correcting parentage relationships in the endangered California Condor: Improving mean kinship estimates for conservation management. Ornithological Applications. https://doi.org/10.1093/ornithapp/duab017
Mucha S, Komen H (2016) Rates of inbreeding and genetic adaptation for populations managed as herds in zoos with a rotational mating system or with optimized contribution of parents. J Anim Breed Genet 133(4):323–332. https://doi.org/10.1111/jbg.12188
Muya SM, Bruford MW, Muigai AWT, Osiemo ZB, Mwachiro E, Okita-Ouma B, Goossens B (2011) Substantial molecular variation and low genetic structure in Kenya’s black rhinoceros: Implications for conservation. Conserv Genet 12(6):1575–1588. https://doi.org/10.1007/s10592-011-0256-3
Okita-Ouma B, van Langevelde F, Heitkönig IMA, Maina P, van Wieren SE, Prins HHT (2021) Relationships of reproductive performance indicators in black rhinoceros (Diceros bicornis michaeli) with plant available moisture, plant available nutrients and woody cover. Afr J Ecol 59(1):2–16. https://doi.org/10.1111/aje.12779
Okita-Ouma, B., Amin, B., & Kock, R. (2007). Conservation and management strategy for the black rhino (Diceros bicornis michaeli) and management guidelines for the white rhino (Ceratotherium simum simum) in Kenya (2007–2011). .
Onley IR, White LC, Moseby KE, Copley P, Cowen S (2022) Disproportionate admixture improves reintroduction outcomes despite the use of low-diversity source populations: population viability analysis for a translocation of the greater stick-nest rat. Anim Conserv. https://doi.org/10.1111/acv.12812
Pacifici M, Rondinini C, Rhodes JR, Burbidge AA, Cristiano A, Watson JEM, Woinarski JCZ, Di Marco M (2020) Global correlates of range contractions and expansions in terrestrial mammals. Nat Commun 11(1):2840. https://doi.org/10.1038/s41467-020-16684-w
Paradis E (2010) pegas: an R package for population genetics with an integrated–modular approach. Bioinformatics 26(3):419–420. https://doi.org/10.1093/bioinformatics/btp696
Patton, F., Campbell, P., & Parfet, E. (2008). Biological management of the high density black rhino population in Solio Game Reserve, central Kenya. Pachyderm, 44, 72–79. https://pachydermjournal.org/index.php/pachyderm/article/view/150
Posit team. (2023). RStudio: Integrated Development Environment for R. http://www.posit.co/
R Development Core Team, R. (2011). R: A Language and Environment for Statistical Computing. In R. D. C. Team (Ed.), R Foundation for Statistical Computing (Vol. 1, Issue 2.11.1, p. 409). R Foundation for Statistical Computing. https://doi.org/10.1007/978-3-540-74686-7
Riester M, Stadler PF, Klemm K (2009) FRANz: Reconstruction of wild multi-generation pedigrees. Bioinformatics 25(16):2134–2139. https://doi.org/10.1093/bioinformatics/btp064
Rohrer GA, Alexander LJ, Keele JW, Smith TP, Beattie CW (1994) A microsatellite linkage map of the porcine genome. Genetics 136(1):231–245. https://doi.org/10.1093/genetics/136.1.231
Rookmaaker LC (1995) Subspecies and ecotypes in the black rhinoceros. Pachyderm 20:39–40
Rookmaaker, L. C., Jones, M. L., Klös, H. G., & Reynolds, R. J. (1998). The rhinoceros in captivity: A list of 2439 rhinoceroses kept from Roman times to 1994. SPB. https://books.google.co.uk/books?id=vDijgNs_7Q0C
Ross CT, Jaeggi AV, Borgerhoff Mulder M, Smith JE, Smith EA, Gavrilets S, Hooper PL (2020) The multinomial index: a robust measure of reproductive skew. Proceedings of the Royal Society B: Biological Sciences 287(1936):20202025. https://doi.org/10.1098/rspb.2020.2025
Sánchez-Barreiro F, De Cahsan B, Westbury MV, Sun X, Margaryan A, Fontsere C, Bruford MW, Russo I-RM, Kalthoff DC, Sicheritz-Pontén T, Petersen B, Dalén L, Zhang G, Marquès-Bonet T, Gilbert MTP, Moodley Y (2023) Historic sampling of a vanishing beast: population structure and diversity in the black rhinoceros. Mol Biol Evol. https://doi.org/10.1093/molbev/msad180
Senn HV, Ghazali M, Kaden J, Barclay D, Harrower B, Campbell RD, Macdonald DW, Kitchener AC (2019) Distinguishing the victim from the threat: SNP-based methods reveal the extent of introgressive hybridization between wildcats and domestic cats in Scotland and inform future in situ and ex situ management options for species restoration. Evol Appl 12(3):399–414. https://doi.org/10.1111/eva.12720
Shaw PD, Graham M, Kennedy J, Milne I, Marshall DF (2014) Helium: visualization of large scale plant pedigrees. BMC Bioinformatics 15(1):259. https://doi.org/10.1186/1471-2105-15-259
van Coeverden de Groot, P. J., Putnam, A. S., Erb, P., Scott, C., Melnick, D., O’Ryan, C., & Boag, P. T. (2011) Conservation genetics of the black rhinoceros, Diceros bicornis bicornis in Namibia. Conserv Gene 12(3):783–792. https://doi.org/10.1007/s10592-011-0185-1
Węcek K, Hartmann S, Paijmans JLA, Taron U, Xenikoudakis G, Cahill JA, Heintzman PD, Shapiro B, Baryshnikov G, Bunevich AN, Crees JJ, Dobosz R, Manaserian N, Okarma H, Tokarska M, Turvey ST, Wójcik JM, Żyła W, Szymura JM, Barlow A (2016) Complex admixture preceded and followed the extinction of wisent in the wild. Mol Biol Evol. https://doi.org/10.1093/molbev/msw254
Wei X, Zhang J (2018) The optimal mating distance resulting from heterosis and genetic incompatibility. Sci Adv. https://doi.org/10.1126/sciadv.aau5518
Weir BS, Goudet J (2017) A unified characterization of population structure and relatedness. Genetics 206(4):2085–2103. https://doi.org/10.1534/genetics.116.198424
Wellmann R, Hartwig S, Bennewitz J (2012) Optimum contribution selection for conserved populations with historic migration. Genet Sel Evol 44(1):34. https://doi.org/10.1186/1297-9686-44-34
Wilder AP, Navarro AY, King SND, Miller WB, Thomas SM, Steiner CC, Ryder OA, Shier DM (2020) Fitness costs associated with ancestry to isolated populations of an endangered species. Conserv Genet 21(3):589–601. https://doi.org/10.1007/s10592-020-01272-8
Williams SE, Hoffman EA (2009) Minimizing genetic adaptation in captive breeding programs: A review. Biol Cons 142(11):2388–2400. https://doi.org/10.1016/j.biocon.2009.05.034
Williams NF, McRae L, Freeman R, Capdevila P, Clements CF (2021) Scaling the extinction vortex: Body size as a predictor of population dynamics close to extinction events. Ecol Evol 11(11):7069–7079. https://doi.org/10.1002/ece3.7555
Willoughby JR, Fernandez NB, Lamb MC, Ivy JA, Lacy RC, DeWoody JA (2015) The impacts of inbreeding, drift and selection on genetic diversity in captive breeding populations. Mol Ecol 24(1):98–110. https://doi.org/10.1111/mec.13020
Wright BR, Farquharson KA, McLennan EA, Belov K, Hogg CJ, Grueber CE (2020) A demonstration of conservation genomics for threatened species management. Mol Ecol Resour 20(6):1526–1541. https://doi.org/10.1111/1755-0998.13211
Acknowledgements
We would like to thank all participating members of the Black rhino European Association of Zoos and Aquaria (EAZA) Endangered species Programme (EEP), particularly the keepers, veterinarians and curators who provided samples for us, as well as the managers and rangers on the South African reserve, without whom this work could not have taken place. This work was funded by a UK Natural Environment Research Council (NERC) PhD studentship with Chester Zoo as a CASE partner, and the NERC Environmental Omics Facility (NEOF). Lab work was performed at the NEOF Visitor Facility at the University of Sheffield.
Funding
This article was funded by Natural Environment Research Council, NERC Biomolecular Analysis Facility, Chester Zoo.
Author information
Authors and Affiliations
Contributions
F.E., S.S., M.P and C.W. conceived the study. F.E. performed the laboratory work and analysed the data with the assistance of D.D. and G.J. F.E. interpreted the data with contributions from S.S. and C.W. P.K. collected and curated observational data. P.K., J.H., T.K and J.H. provided samples. F.E. wrote the first draft of the paper with input from all other authors.
Corresponding author
Ethics declarations
In compliance with the Nagoya agreement, samples from South Africa were imported with prior informed consent and an access and benefit sharing agreement with permit number: AWB No. 7220347514.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Elsner-Gearing, F., Kretzschmar, P., Shultz, S. et al. Admixture and reproductive skew shape the conservation value of ex situ populations of the Critically Endangered eastern black rhino. Conserv Genet 25, 897–910 (2024). https://doi.org/10.1007/s10592-024-01611-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-024-01611-z