Abstract
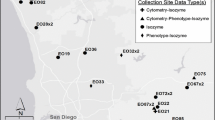
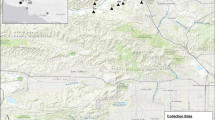
Understanding genetic structure in rare plant populations is essential to making informed decisions for recovery actions, particularly in species capable of clonal reproduction. Here, we present three case studies using microsatellites to assess clonal structure in rare plants: Northern Oconee bells (S. brevistyla (P.A. Davies) Gaddy) and Southern Oconee bells (Shortia galacifolia Torr & Gray) (Diapensiaceae); and bunched arrowhead (Sagittaria fasciculata E.O. Beal (Alismataceae)). We used six loci to genotype Shortia brevistyla (n = 62 ramets; three sites) and S. galacifolia (n = 111 ramets; seven sites) and five loci in Sagittaria fasciculata (n = 162 ramets; eight sites). Ramets were systematically mapped and sampled to allow for clonal assignment in a spatial context. All ramets for S. brevistyla were genetically identical across all loci, while S. galacifolia exhibited variation consistent with a mixed reproductive strategy. Sagittaria fasciculata also exhibited a mixed reproductive strategy with emphasis on clonality. Our data indicate that stem counts are not effective measures for recovery assessment in these species, and a more complex demographic monitoring protocol should be developed. Additional implications for conservation of these species are discussed, including a consideration for federal listing for Shortia brevistyla.





Similar content being viewed by others
Data availability
The data generated in the present study are available from the corresponding author upon request.
References
Araki KS, Shimatani IK, Ohara M (2023) Genet dynamics and its variation among genets of a clonal plant Convallaria keiskei. Oikos, 2023, e09367
Arnaud-Haond S, Duarte CM, Alberto F, Serrao EA (2007) Standardizing methods to address clonality in population studies. Mol Ecol 16:5115–5139
Barrett SC, Colautti RI, Eckert CG (2008) Plant reproductive systems and evolution during biological invasion. Mol Ecol 17:373–383
Brzyski JR, Stieha CR, Nicholas Mcletchie D (2018) The impact of asexual and sexual reproduction in spatial genetic structure within and between populations of the dioecious plant Marchantia inflexa (Marchantiaceae). Ann Botany 122:993–1003
Calevo J, Gargiulo R, Bersweden L, Viruel J, González-Montelongo C, Rebbas K, Boutabia L, Fay MF (2021) Molecular evidence of species-and subspecies-level distinctions in the rare Orchis patens s.l. and implications for conservation. Biodivers Conserv 30:1293–1314
Callaghan TV, Carlsson BÅ, Jónsdóttir IS, Svensson BM, Jonasson S (1992) Clonal plants and environmental change: introduction to the proceedings and Summary. Oikos 63:341–347
Carrillo Angeles IG, Golubov J, Milligan BG, Mandujano MC (2011) Spatial distribution pattern of a clonal species: effects of differential production of clonal and sexual offspring. Evol Ecol 25:1357–1383
Charpentier A, Mesleard F, Thompson J (1998) The effects of rhizome severing on the clonal growth and clonal architecture of Scirpus maritimus. Oikos, pp 107–116
Coates DJ, Byrne M, Moritz C (2018) Genetic diversity and conservation units: dealing with the species-population continuum in the age of genomics. Front Ecol Evol 6:165
Cohen JI, Ruane LG (2022) Conservation genetics of Phlox hirsuta, a serpentine endemic. Conservation Genetics, pp 1–16
da Cunha NL, Xue H, Wright SI, Barrett SC (2022) Genetic variation and clonal diversity in floating aquatic plants: comparative genomic analysis of water hyacinth species in their native range. Mol Ecol 31:5307–5325
Davies P (1952) Geographical variations in Shortia galacifolia. Rhodora 54:121–124
Davies P (1960) Pollination and seed production in Shortia galacifolia. Castanea 25:89–96
De Witte LC, Stöcklin J (2010) Longevity of clonal plants: why it matters and how to measure it. Ann Botany 106:859–870
Delnevo N, Piotti A, van Etten EJ, Stock WD, Byrne M (2019) Isolation, characterization, and cross-amplification of 20 microsatellite markers for Conospermum Undulatum (Proteaceae). Appl Plant Sci 7:e11283
Dorken ME, Eckert CG (2001) Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). J Ecol, 339–350
Douhovnikoff V, Dodd RS (2015) Epigenetics: a potential mechanism for clonal plant success. Plant Ecol 216:227–233
Dripps W, Lewis GP, Baxter R, Andersen CB (2013) Hydrogeochemical characterization of headwater seepages inhabited by the endangered bunched arrowhead (Sagittaria fasciculata) in the upper Piedmont of South Carolina. Southeast Nat 12:619–637
Dunn BA, Jones SM (1979) Geographical distribution of Shortia galacifolia in Oconee and Pickens Counties, South Carolina. J Elisha Mitchell Sci Soc 95:32–41
Eckert CG, Dorken ME, Barrett SC (2016) Ecological and evolutionary consequences of sexual and clonal reproduction in aquatic plants. Aquat Bot 135:46–61
Ellstrand NC, Roose ML (1987) Patterns of genotypic diversity in clonal plant species. Am J Bot 74:123–131
Faircloth BC (2008) MSATCOMMANDER: detection of microsatellite repeat arrays and automated, locus-specific primer design. Mol Ecol Resour 8:92–94
Fehlberg SD (2017) Knowledge of genetic diversity informs conservation of a rare, clonal wetland plant Lilaeopsis schaffneriana subsp. recurva (Apiaceae). Journal of the Botanical Research Institute of Texas, pp 499–510
Gaddy L, Carter T, Ely B, Sakaguchi S, Matsuo A, Suyama Y (2019) Shortia brevistyla comb. Et stat. nov., (Diapensiaceae), a narrow endemic from the headwaters of the Catawba River in North Carolina, USA. Phytologia 101:113–119
Gargiulo R, Ilves A, Kaart T, Fay MF, Kull T (2018) High genetic diversity in a threatened clonal species, Cypripedium calceolus (Orchidaceae), enables long-term stability of the species in different biogeographical regions in Estonia. Bot J Linn Soc 186:560–571
Gitzendanner MA, Weekley CW, Germain-Aubrey CC, Soltis DE, Soltis PS (2012) Microsatellite evidence for high clonality and limited genetic diversity in Ziziphus celata (Rhamnaceae), an endangered, self-incompatible shrub endemic to the Lake Wales Ridge, Florida, USA. Conserv Genet 13:223–234
Godoy FMR, Lenzi M, Ferreira BHDS, Silva LVD, Zanella CM, Paggi GM (2018) High genetic diversity and moderate genetic structure in the self-incompatible, clonal Bromelia hieronymi (Bromeliaceae). Bot J Linn Soc 187:672–688
Guo L, Plunkert M, Luo X, Liu Z (2021) Developmental regulation of stolon and rhizome. Curr Opin Plant Biol 59:101970
Han Y-Q, Wang L-G, You W-H, Yu H-H, Xiao K-Y, Wu Z-H (2018) Flooding interacting with clonal fragmentation affects the survival and growth of a key floodplain submerged macrophyte. Hydrobiologia 806:67–75
Herben T, Klimešová J (2020) Evolution of clonal growth forms in angiosperms. New Phytol 225:999–1010
Hodel RG, Segovia-Salcedo MC, Landis JB, Crowl AA, Sun M, Liu X, Gitzendanner MA, Douglas NA, Germain‐Aubrey CC, Chen S (2016) The report of my death was an exaggeration: a review for researchers using microsatellites in the 21st century. Appl Plant Sci 4:1600025
Honnay O, Jacquemyn H (2007) Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conserv Biol 21:823–831
Jacquemyn H, Brys R, Honnay O, Hermy M, Roldán-Ruiz I (2005) Local forest environment largely affects below-ground growth, clonal diversity and fine‐scale spatial genetic structure in the temperate deciduous forest herb Paris Quadrifolia. Mol Ecol 14:4479–4488
Jones SM, Augspurger MK (1988) Seed germination and phenology of Shortia galacifolia T.&G. Castanea, pp 140–148. Diapensiaceae
Keener BR (2005) Molecular systematics and revision of the aquatic monocot genus Sagittaria (Alismataceae). Dissertation, The University of Alabama
Kocot D, Sitek E, Nowak B, Kołton A, Stachurska-Swakoń A, Towpasz K (2022) The effectiveness of the sexual reproduction in selected clonal and nonclonal species of the genus Ranunculus, vol 11. Biology, p 85
Lambertini C, Riis T, Olesen B, Clayton JS, Sorrell BK, Brix H (2010) Genetic diversity in three invasive clonal aquatic species in New Zealand. BMC Genomic Data 11:1–18
Larkin PD, Hamilton AM, Lopez AI, Rubiano-Rincon S (2020) How clone can you go? Seedbank density and a multiscale assessment of genotypic diversity in the seagrass Halodule wrightii. Aquat Bot 163:103207
LeGrand H, Sorrie B, Howard T (2023) Vascular plants of North Carolina. North Carolina Biodiversity Project and North Carolina State Parks, Raleigh, NC
Lin CH, Miriti MN, Goodell K (2016) Demographic consequences of greater clonal than sexual reproduction in Dicentra canadensis. Ecol Evol 6:3871–3883
Manry KF (2002) Breeding system and timing of Inbreeding depression in the rare plant Shortia galacifolia (Diapensiaceae). Thesis, Clemson University
Maschinski J, Albrecht MA (2017) Center for Plant Conservation’s best practice guidelines for the reintroduction of rare plants. Plant Divers 39:390–395
Meirmans PG, Van Tienderen PH (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Morris AB, Scalf C, Burleyson A, Johnson LT, Trostel K (2016) Development and characterization of microsatellite primers in the federally endangered Astragalus bibullatus (Fabaceae). Applications in Plant Sciences, 4, 1500126
NatureServe (2023) NatureServe Network Biodiversity Location Data accessed through NatureServe Explorer [web application]. NatureServe, Arlington, Virginia
Newberry G (1991) Factors affecting the survival of the rare plant, Sagittaria fasciculata EO Beal (Alismataceae). Castanea, pp 59–64
Newberry G (2000a) Assessment of the sagittaria fasciculata (bunched arrowhead) populations in North and South Carolina. Oct 20, 2000. 130
Newberry G (2000b) Third year report on the biological monitoring ofS agittaria fasciculata. Dec, 1986. 49
Nguyen TM, Vu DD, Nguyen DM, Dang HP, Phan LK, Bui PX (2020) Microsatellite analysis reveals genetic diversity of the endangered species Dipterocarpus Dyeri. J for Res 25:198–201
Nguyen X-V, Nguyen-Nhat N-T, Nguyen X-T, Dao V-H, McDermid KJ, Papenbrock J (2022) Microsatellite-based analysis of the genetic diversity and population structure of the seagrass species Thalassia hemprichii from southern Viet Nam. Aquat Bot 178:103497
Niklas KJ, Cobb ED (2017) The evolutionary ecology (evo-eco) of plant asexual reproduction. Evol Ecol 31:317–332
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Oostermeijer J, Luijten S, Den Nijs J (2003) Integrating demographic and genetic approaches in plant conservation. Biol Conserv 113:389–398
Ouborg N, Vergeer P, Mix C (2006) The rough edges of the conservation genetics paradigm for plants. J Ecol 94:1233–1248
Parveen S, Singh N, Adit A, Kumaria S, Tandon R, Agarwal M, Jagannath A, Goel S (2022) Contrasting reproductive strategies of two Nymphaea species affect existing natural genetic diversity as assessed by microsatellite markers: implications for conservation and wetlands restoration. Front Plant Sci, 13
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28:2537–2539
Potter D, Bartosh H, Dangl G, Yang J, Bittman R, Preece J (2018) Clarifying the conservation status of Northern California black walnut (Juglans hindsii) using microsatellite markers. Madroño 65:131–140
Rozen S, Skaletsky H (1999) Primer3 on the WWW for general users and for biologist programmers. Bioinf Methods Protocols, 365–386
Schoen DJ, Schultz ST (2019) Somatic mutation and evolution in plants. Annu Rev Ecol Evol Syst 50:49–73
Schreiber SJ, Yamamichi M, Strauss SY (2019) When rarity has costs: coexistence under positive frequency-dependence and environmental stochasticity. Ecology 100:e02664
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234
Selkoe KA, Toonen RJ (2006) Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol Lett 9:615–629
Silvertown J (2008) The evolutionary maintenance of sexual reproduction: evidence from the ecological distribution of asexual reproduction in clonal plants. Int J Plant Sci 169:157–168
Spielman D, Brook BW, Frankham R (2004) Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci 101:15261–15264
STATEMENTS & DECLARATIONS
Taft HR, McCoskey DN, Miller JM, Pearson SK, Coleman MA, Fletcher NK, Mittan CS, Meek MH, Barbosa S (2020) Research–management partnerships: an opportunity to integrate genetics in conservation actions. Conserv Sci Pract, 2, e218
Tepedino V (2012) Overestimating population sizes of rare clonal plants. Conserv Biol 26:945–947
USFWS (2016) USFWS Species Status Assessment Framework: an integrated analytical framework for conservation. In: Version 34 dated August 2016
USFWS (2020) Bunched arrowhead (Sagittaria fasciculata) 5-year review: summary and evaluation. In: US Fish and Wildlife Service, pp. 1–22, Asheville, NC
USFWS (1983) Bunched arrowhead recovery plan. In: US Fish and Wildlife Service, pp. 1–37, Atlanta, Georgia
USGS (2019) National Hydrography Dataset (ver. USGS National Hydrography Dataset Best Resolution (NHD) for Hydrologic Unit (HU) 4–2001 (published 20191002)). https://www.usgs.gov/national-hydrography/access-national-hydrography-products. Accessed June 28, 2023
Vallejo-Marín M, Dorken ME, Barrett SC (2010) The ecological and evolutionary consequences of clonality for plant mating. Annu Rev Ecol Evol Syst 41:193–213
Vivian VE (1967) Shortia galacifolia: its life history and microclimatic requirements. Bull Torrey Bot Club 94:369–387
Wang Z, Xie L, Prather CM, Guo H, Han G, Ma C (2018) What drives the shift between sexual and clonal reproduction of Caragana stenophylla along a climatic aridity gradient? BMC Plant Biol 18:1–10
Ward S (2006) Genetic analysis of invasive plant populations at different spatial scales. Biol Invasions 8:541–552
Weakley AS (2022) Flora of the southeastern United States. In: University of North Carolina Herbarium, North Carolina Botanical Garden
Willi Y, Kristensen TN, Sgrò CM, Weeks AR, Ørsted M, Hoffmann AA (2022) Conservation genetics as a management tool: The five best-supported paradigms to assist the management of threatened species. Proceedings of the National Academy of Sciences, 119, e2105076119
Zahner R, Jones SM (1983) Resolving the type location for Shortia galacifolia T.&G, vol 48. Castanea, pp 163–173
Zane L, Bargelloni L, Patarnello T (2002) Strategies for microsatellite isolation: a review. Mol Ecol 11:1–16
Zhang Y, Zhang D (2007) Asexual and sexual reproductive strategies in clonal plants. Front Biology China 2:256–262
Zhang H, Chen SC, Bonser SP, Hitchcock T, Moles AT (2023) Factors that shape large-scale gradients in clonality. Journal of Biogeography
Zheng Z, Hu H, Lei W, Zhang J, Zhu M, Li Y, Zhang X, Ma J, Wan D, Ma T (2022) Somatic mutations during rapid clonal domestication of Populus alba var. pyramidalis. Evol Appl 15:1875–1887
Acknowledgements
This research was funded by the U.S. Fish and Wildlife Service (Grant No. F22AP02689-00), the South Carolina Native Plant Society Upstate Chapter, Friends of Jocassee, the Furman University Office of Undergraduate Research (Summer Research Fellow Awards to LE, JBF, CP, LS, NT, and LV), the Furman University Research and Professional Growth Committee (award to ABM), and the Sewanee Herbarium (support to SF).We thank the following people for assisting with permitting, site access, field work, and helpful discussions regarding these projects: Sam Tessel, Keith Bradley, and Austen Attaway (SCDNR); Brenda Wichmann (NCDCR); Katherine Culatta, Lesley Starke, and Kate Loughran (NCPCP); Wes Knapp (NatureServe); Stacy Scherman (SC Parks); Alan Stuart (Duke Energy); and Kevin Evans and Rowdy Harris (Devils Fork SP). Special thanks to Dan Whitten and Kay Wade for their invaluable support and assistance with work regarding the Shortia complex and Steve Bogdanowicz (Cornell University) for microsatellite development and troubleshooting. We thank Min-Ken Liao for sharing her unpublished ISSR data for Sagittaria fasciculata. We thank Sam Tessel, Keith Bradley, Katherine Culatta, Kate Loughran, Jon Evans (Sewanee – University of the South), Natali Ramirez-Bullon (USFWS Asheville Field Office), and two anonymous reviewers for feedback on previous versions of this manuscript.
Funding
This research was funded by the U.S. Fish and Wildlife Service (Grant No. F22AP02689-00), the South Carolina Native Plant Society Upstate Chapter, Friends of Jocassee, the Furman University Office of Undergraduate Research (Summer Research Fellow Awards to LE, JBF, CP, LS, NT, and LV), the Furman University Research and Professional Growth Committee (award to ABM), and the Sewanee Herbarium (support to SJF).
Author information
Authors and Affiliations
Contributions
Ashley B. Morris conceptualized the project, acquired funding, and supervised all research efforts. Skyler Fox and Ashley B. Morris were responsible for project administration. All authors collected leaf material in the field. Skyler Fox, Lauren Eberth, J. Banks Floyd, Calla Pederson, Lily Stafford, Nora Tillmanns, and Lo Vodo performed lab work. Skyler Fox curated data for the project. Skyler Fox, Lauren Eberth, J. Banks Floyd, Calla Pederson, Lily Stafford, Nora Tillmanns, and Ashley B. Morris analyzed data and contributed to drafts of the manuscript. All authors read and approved the final manuscript. Parts of this study were completed by Lauren Eberth, J. Banks Floyd, Calla Pederson, Lily Stafford, and Nora Tillmanns as part of the requirements for undergraduate senior thesis research in the Department of Biology at Furman University.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fox, S., Eberth, L., Floyd, J.B. et al. The importance of understanding clonal structure for species listing and recovery: case studies from the rare oconee bells (Shortia brevistyla and Shortia galacifolia; Diapensiaceae) and the federally endangered bunched arrowhead (Sagittaria fasciculata; Alismataceae). Conserv Genet (2024). https://doi.org/10.1007/s10592-024-01608-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10592-024-01608-8