Abstract
The harbour porpoise (Phocoena phocoena), a highly mobile cetacean species of the Northern Hemisphere, inhabits basins that vary broadly in salinity, temperature, and food availability; such variation can drive divergent adaptation among local populations. To shed light on range-wide population structure and local adaptation, we generated ddRAD sequencing data spanning the entire North Atlantic and the Baltic Sea, as well as the Black Sea as an outgroup, and mapped this data to the high-quality draft genome of the species. We identified 11,978 genome-wide SNPs from 150 individuals, which we used for population genetic inferences. Our results support genetic differentiation between North Atlantic and Baltic Sea populations, with Kattegat as a transition zone. Across the North Atlantic the population differentiation is subtle from west to east, congruent with an isolation-by-distance pattern, but indicates a separation of southern North Sea harbour porpoises. We identified genomic outlier regions, i.e., scaffold regions where SNPs with high FST across North Atlantic populations co-occur. Together with the draft genome annotation, these regions could point towards candidate genes for differential local adaptation processes among populations. Furthermore, they enable the development of a SNP panel for routine population assignment which will be useful in a conservation and management context. We identified six outlier loci putatively under positive selection, based on the population structure inferred from the complete SNP set. Our study highlights the value of genome resources in conservation and management and provides a crucial additional resource for the study of harbour porpoise evolution and phylogeny.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The harbour porpoise (Phocoena phocoena) is a small cetacean species occurring in coastal habitats in the Northern Hemisphere (Fig. 1; Fontaine et al. 2014, 2017; Galatius et al. 2012; Lah et al. 2016). It comprises five described subspecies and potentially additional not yet formally described ones, i.e. P. p. vomerina (North Pacific), unnamed (Pacific), P. p. meridionalis (South Atlantic Ocean and Iberian Sea), P. p. relicta (Black Sea) and P. p. phocoena (North Atlantic), of which some display locally different traits in morphology and behaviour (ASCOBANS 2021; Carlén et al. 2018; Fontaine et al. 2017; Galatius et al. 2012; NAMMCO 2019). The North Atlantic harbour porpoise (P. p. phocoena) is distributed from Canada and Greenland to the North and the Baltic Sea. Within these regions, the sea basins exhibit a wide range of temperature, currents, salinity and other water conditions representing a variety of marine habitats, potentially posing different selective pressures to the species. The porpoises across the North Atlantic show variation in diet (Aarefjord et al. 1995; Hammond et al. 2013; Víkingsson et al. 2003), differences in habitat use and activity patterns (Nuuttila et al. 2017), fine scale morphological differences (Galatius et al. 2012; Härkönen et al. 2013; Viaud-Martínez et al. 2007) and significant isolation-by-distance (Lah et al. 2016). Based on these differences, local ecotypes have been postulated within the North Atlantic (Fontaine et al. 2010, 2017; Hammond et al. 2020; Olsen et al. 2022; Santos et al. 2004) and the Baltic Sea (Celemin et al. 2023; Galatius et al. 2012; Lah et al. 2016). The Baltic Sea was colonized from the North Sea during the end of the last glacial period (Sommer et al. 2008). Within the Baltic Sea, several small basins are separated by underwater ridges and are further fragmented by multiple small islands and headlands. Today, the Baltic Sea differs in temperature and salinity from the North Sea/North Atlantic, as it has transitioned from freshwater to brackish and marine water since its formation in the Pleistocene (~ 15 kyr ago) (Paasche et al. 2015; Varjopuro et al. 2014). This could have promoted the divergence of harbour porpoises in the Baltic Sea from populations of the North Sea/North Atlantic (Celemin et al. 2023; Lah et al. 2016; Wiemann et al. 2010).
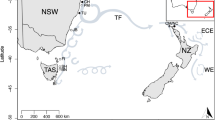
Distribution map of the harbour porpoise in the North Atlantic and adjacent waters. Occurrence of the species is indicated by orange colouring. Black circle sizes indicate the relative number of samples per location (Canada (CA), Iceland (ICE), North Sea (NOS), Skagerrak (SKA), Kattegat (KAT), Belt Sea (BES), Inner Baltic Sea (IBS), and as outgroup Western Black Sea (WBS)). Map taken from IUCN Red List (IUCN (International Union for Conservation of Nature) 2012. Phocoena phocoena. The IUCN Red List of Threatened Species. Version 2019-3)
Across its North Atlantic distribution, the abundance of the harbour porpoise varies greatly. The European Atlantic Shelf is estimated to be inhabited by ~ 424,000 individuals (Hammond et al. 2018), with 43,000 animals in Icelandic coastal waters (Gilles et al. 2020) and approximately 14,400 animals estimated in the Belt Sea including Kattegat and the SW Baltic Sea (Gilles et al. 2023). The Proper Baltic porpoise population has much lower estimates (~ 500 animals in summer) and is considered critically endangered (Benke et al. 2014; Carlén et al. 2018; Carlström et al. 2023; Hammond et al. 2020; SAMBAH 2016; Scheidat et al. 2008).
With their ocean-wide distribution, cetaceans encounter a number of different threats from humans utilizing the marine habitat (McCauley et al. 2015). Humans contribute to marine debris (Siebert et al. 2001; Unger et al. 2017), cause bycatch (Kesselring et al. 2017; Omeyer et al. 2020) and noise pollution, especially in forms of offshore work and shipping routes (Dyndo et al. 2015; Farmer et al. 2018; Guzman et al. 2013; Holt et al. 2017; Rolland et al. 2012; Wisniewska et al. 2018). These disturbances could impose barrier effects, leading to decrease or fragmentation of populations (Dungan et al. 2016; Fontaine et al. 2007).
Harbour porpoises have a relatively high nutrient demand because of their small size, their reproductive cycle and the fact that they lactate while reproducing and often have a calf every year (Kesselring et al. 2017; Wisniewska et al. 2016). They mainly inhabit coastal areas, which increases their risk of negative exposure to human threats (Fontaine et al. 2010).
To assess the status of harbour porpoise populations, different techniques have been applied: acoustic monitoring, tagging and aerial surveys to assess distribution, abundance and habitat use (Lonergan et al. 2011; Pike et al. 2020; SAMBAH 2016; Sveegaard et al. 2015), as well as regular autopsies on stranded and bycaught specimens to provide information on health, pathology, age structure, and reproductive status (IJsseldijk et al. 2020; Kesselring et al. 2017; Siebert et al. 2001, 2006, 2020). In the context of assessment, it is particularly important to understand the genetic structure and divergence among populations (McMahon et al. 2014; Waples et al. 2018). Cetaceans are highly mobile in a seemingly continuous habitat, yet they follow abundance of their food, and they may be constrained by further specific habitat characteristics (Baker et al. 2013; Jefferson 2014; Van Cise et al. 2019). Hence, genetic structure may be difficult to understand and subtle structuring may be overlooked easily (Leslie and Morin 2016).
Cetacean sampling is often opportunistic relying on bycaught or stranded specimens. Sample material may hence often be degraded and not all sampling regions are equally represented (ASCOBANS 2002; Durban et al. 2005; Luca et al. 2009; Pierce et al. 2007). When relying on opportunistic sampling, it is even more important to have markers that show high resolution even with little genetic material or a low number of samples per geographic region. High-quality genomes for whales and dolphins have only recently been generated and made accessible (e.g. Autenrieth et al. 2018; Gao et al. 2023; Neely et al. 2018; Yu et al. 2022; Yuan et al. 2018; Zhou et al. 2018). Due to the vastly improved resolution in comparison to studies on single loci, even subtle population differentiation can be detected (Cammen et al. 2016; Çilingir et al. 2022; de Greef et al. 2022; Gallego-García et al. 2021). In recent years genome wide single nucleotide polymorphisms (SNPs) have become valuable markers to identify population structure and boundaries (Çilingir et al. 2022; Liu et al. 2005; Morin et al. 2009; Santure et al. 2010). Restriction site associated DNA sequencing techniques (RADseq, ddRAD), which allow for SNP discovery and genotyping, have become widely used approaches for population genomic analyses in non-model organisms, such as cetaceans (Attard et al. 2018; Cammen et al. 2016; Carroll et al. 2016; Davey et al. 2011; Reeves et al. 2022; Viricel et al. 2014).
The aim of this study is to provide a high-resolution genetic marker system, which allows for detection of even subtle population structure over the North Atlantic distribution range of harbour porpoise. We therefore extend on a previous study (Lah et al. 2016) using ddRAD sequencing, by increasing the sample size and geographic coverage. Furthermore, we use the published genome of the harbour porpoise (Autenrieth et al. 2018) as a mapping reference to increase the resolution of our newly obtained genome-wide SNPs. With this improved data set, we aim to resolve the population structure of harbour porpoise across the entire North Atlantic, as well as within the Baltic Sea, and further provide an informative set of SNPs for the investigation of local population differentiation and adaptation with possible implications for conservation.
Materials and methods
Sampling locations
We focused this study on the North Atlantic distribution range of the species. We took 151 samples (Supplemental Table 1) from eight different ‘sampling areas’: Canada (CA), Iceland (ICE), North Sea (NOS), Skagerrak (SKA), Kattegat (KAT), Belt Sea (BES), Inner Baltic Sea (IBS) and - as an outgroup – Western Black Sea (WBS) (Fig. 1). The boundaries of these areas follow a previous study and are based on definitions by the International Council for the Exploration of the Sea, ICES (Wiemann et al. 2010). Additionally, we defined three ‘regions’, based on the main sea basins: West North Atlantic (WNA, including CA + ICE), East North Atlantic/North Sea (ENA, including NOS + SKA) and Baltic Sea (BALT, including BES + IBS). These regions exclude the Black Sea as an outgroup and Kattegat as a transition zone (Lah et al. 2016; Rosel et al. 1999; Wiemann et al. 2010). All sampling was performed on dead by-caught or stranded carcasses, and no live harbour porpoises have been targeted for this study. The sampling was performed by persons authorised by the respective national authorities.
Wet lab procedure
Total genomic DNA was extracted from tissue samples (skin or muscle) stored at -20 °C (in ethanol or frozen) with the NucleoSpin Tissue Kit (Macherey-Nagel, Germany) following the manufacturer’s protocol. Canadian samples were extracted with the DNeasy Blood and Tissue kit (QIAGEN GmbH, Germany). To assess sample quality and quantity, DNA concentration was measured on a NanoDrop 1000 (Thermo Scientific, USA) and an Agilent 2200 TapeStation (Genomic ScreenTape System; Agilent Technologies, USA). Using traditional Sanger sequencing, we genotyped each sample’s mitochondrial control region as an independently inherited marker previously shown to be informative about population structure (Supplemental Table 1). Polymerase Chain Reactions (PCR) were performed on a BiometraT3000 thermocycler, using the primers ProL and DLH and following previously established protocols (Tiedemann et al. 1996; Wiemann et al. 2010). Using the Applied Biosystems BigDye chemistry, sequencing was performed on a 3130xl Genetic Analyzer (Applied Biosystems).
For genomic library preparation, we chose a modified ddRAD sequencing method, using the restriction enzymes PstI and MspI. All samples were sequenced on an Illumina HiSeq 2000. Paired end reads with a length of 100 bp were sequenced with a sequencing amount of 200 million read pairs. The digestion, library preparation and sequencing were carried out by a commercial sequencing company (LGC Genomics, Berlin), as detailed in Lah et al. 2016.
ddRAD-seq data analyses and SNP calling
LGC Genomics provided reads with clipped adapters and quality filtering. These reads did not contain any missing data, were trimmed at the 3’end and had a minimum average Phred quality score of 20 over a window of 10 bases, as well as a minimum length of 64 bp. We mapped these reads against the genome of the harbour porpoise (Autenrieth et al. 2018) using bwa mem vs.0.7.17-r1188 (Li and Durbin 2010) with a Phred score of 30. Since the reference genome does not contain mtDNA sequences, we did not specifically filter for mtDNA reads. Since the X-chromosome is not present as single scaffolds/not unambiguously identified, we did not map ddRAD reads specifically to the X chromosome. Duplicates were removed and all bam files were indexed with samtools vs.1.3.1 (Li et al. 2009).
Several bioinformatics tools and pipelines for RADseq analyses were developed in recent years, based on different algorithms and considering different models for variant calling. They result in different rates of false discovery and information delivered when calling variants and SNPs (Cornish and Guda 2015; Korneliussen et al. 2014; Maruki and Lynch 2017; Mielczarek and Szyda 2016; Rochette et al. 2019; Shafer et al. 2017). Therefore, to have the most confident SNPs and as recommended by some review papers (Kumaran et al. 2019; Shafer et al. 2017), we decided to combine different commonly applied pipelines and kept only SNPs identified by all of them for downstream analysis. Specifically, we used three different programs with four different models: bcftools vs.1.9, angsd vs. 0.929 (models gatk and samtools; Korneliussen et al. 2014), stacks (gstacks vs2.2 and populations vs2.2; Rochette et al. 2019). bcftools was used with minimum mapping and minimum base quality set to 20 and using the model for multiallelic and rare-variant calling with an expected substitution rate of 1e− 6. angsd provides different models for estimating the genotype likelihood, of which we applied the more widely used models ‘gatk’ and ‘samtools’ (see Korneliussen et al. 2014 for more details on the calculations). In both runs, we let the program calculate the per-site allele frequency and posterior probability, major and minor alleles were inferred, and p-value of site was set to be 10e− 6. gstacks was run with standard settings, including the restriction that only one SNP per stack was called to reduce the likelihood of linked SNPs, followed by populations (also with standard settings), which was used to extract called SNP positions and create a vcf file for downstream analysis.
Using a custom R script, we looked for overlap in the results of the chosen pipelines (bcftools, stacks, angsd) and extracted only those SNPs supported by all of them. The overlapping SNPs were filtered for a quality threshold of 20, minimum depth of 100 and mean genotype depth above 5 using bctools vs.1.9 and vcftools vs0.1.15. Only one sample (PPC30-Pool2-102 from Canada) had a high amount of missing data (97%) and was excluded; for the remaining samples (n = 150), missing data were below 5%. We then created two datasets, one including the Black Sea (150 samples, referred to as ‘setA’ from here on), which then was filtered for a minor allele frequency of 2% for all samples, and a second dataset excluding the Black Sea (145 samples, referred to as ‘setB’ from here on) which we filtered for an allele frequency of at least 5%. As a final filter step, we kept only SNPs occurring in 100% of samples for both sets (setA and setB). vcftools was used to calculate SNP-wise FST between sampling areas (8 for setA, 7 for setB) as well as among the defined regions WNA, ENA and BALT. These FST were then visualized with the package ggplot2 in R as function of the scaffold wide SNP distribution per scaffold, where the positions of the SNPs along the scaffolds were illustrated. Using the python script popgen, we calculated the Fst for sliding windows along the scaffolds with a 500 kb window size, a minimum of 5 SNP sites occurring and no overlap between windows allowed.
SNP-based population statistics and differentiation analyses
Using the program arlequin 3.5.2.2. (Excoffier and Lischer 2010) vs2.2, we calculated diversity indices, including nucleotide diversity and expected heterozygosity. Using the tool vcftools, we calculated observed heterozygosity for both SNP sets, applying sampling areas as units. Genetic diversity differences among samples were analysed using a hierarchical AMOVA in arlequin. Sequential Bonferroni tests (Sokal and Rohlf 2012) were performed to correct for multiple comparisons of pairwise FST, which were also calculated in arlequin.
We used discriminant analysis of principal components (DAPC) and admixture to estimate the number of clusters across all sampling areas without prior region/population assignment. To maximize the discrimination between potential clusters, a DAPC separates the variance of each sample into between and within group components. Clusters are inferred through a discriminant analysis based on the initial principal component analysis of the data. The DAPC, as well as a PCA, AMOVA and pairwise fixation indices FST among sampling regions were calculated and illustrated with R, using the packages adegenet (Jombart 2008), pegas (Paradis 2010), hierfstat (Goudet 2005) and StAMPP (Pembleton et al. 2013). The program admixture (Alexander et al. 2009) was used to infer clusters and genetic identity of each sample to the respective clusters. In admixture, which adopts the likelihood model embedded in the program structure (Pritchard et al. 2000), estimation and evaluation of the best K was performed using cross-validation. admixture computes maximum likelihood estimates in a parametric model for individual ancestry estimates. We used R studio (RStudio Team 2015) for visualization and tableau2019.3.0 to plot the genetic cluster identity of each sample geographically, according to the sample’s coordinates.
Detection of outlier loci
Outlier SNPs were detected using bayescan vs.2.1. (Foll and Gaggiotti 2008). bayescan identifies candidate loci based on differences in allele frequencies between populations by applying a multinomial-Dirichlet model (Foll 2012). To evaluate whether outliers may be indicative of local adaptation, selection was inferred using a logistic regression to divide the FST coefficient into a population specific (β) and a locus specific component (α). We set the posterior odds for the neutral model to 1,000, used 20 pilot runs with a thinning set to 5,000 and burn-in of 50,000, followed by a total of 100,000 iterations. A q-value, the bayescan FDR (false discovery rate) analogue to the p-value, of 10% was applied. Using plink vs1.07 (Purcell et al. 2007), we calculated the SNP-wise allele frequency and extracted outlier SNPs (based on the α (> 10) and FST value (> 0.05) calculated by Bayescan). This analysis was performed twice, i.e., comparing the allele frequencies between (1) the different sampling areas and (2) the different regions. Using custom scripts, we then compared the outlier SNPs to the draft annotation of the reference genome (Autenrieth et al. 2018) to see if SNPs are localized in annotated regions.
Mitochondrial control region data analysis
Mitochondrial control region sequences were aligned in MEGA vs.10.1.7. (Kumar et al. 2018) and haplotypes were defined based on sequence comparisons with previously published haplotypes (Lah et al. 2016; Wiemann et al. 2010). Using the program arlequin, we calculated genetic diversity indices, pairwise FST and performed an AMOVA. A haplotype network based on the mitochondrial control region was created using the program popart vs.1.7. (Leigh and Bryant 2015). Additionally, we calculated the haplotype frequency per sampling area and region.
Results
DNA quality and sequencing output
The number of raw reads per individual was on average 2,220,724 with the lowest number being 1,224,194 and the highest 3,137,076. After mapping the reads of each sample to the genome, we used the program samtools vs1.3.1. to calculate mapping statistics (Table 1). We observed an average mapping quality per sample of 36.84 and a mean number of mapped reads per sample of 1,681,717.33 reads (Table 1). The information for all samples is provided in the Supplemental Table 1.
SNP calling, filtering and distribution
The number of overall called SNPs varied depending on the pipeline used: bcftools (985,537), angsd (gatk: 2,692,730, samtools: 2,003,753) and stacks (330,307). The number of SNPs consistently called across all four algorithms and retained for all further analyses was 269,500 (Supplemental Fig. 1). After all filter steps (minimum quality of 20, minimum depth of 100 and mean genotype depth above 5) were performed for SNPs shared among all models, the dataset encompassed a total of 52,175 SNPs. For SNP setA (Black Sea included), we retained only those SNPs found in 100% of the 150 samples, resulting in a final dataset of 26,320 SNPs. For SNP setB (Black Sea excluded), 24,705 and 11,978 SNPs were present in at least 97% and 100% of the samples, respectively. Given the relatively high number of retained SNPs, and to facilitate direct comparison with SNP setA, we also decided to continue with the 100% filter for downstream analyses of setB. Finally, in mapping the ddRAD reads to the genome, we found that the SNPs were more or less evenly distributed across scaffolds (Supplemental Fig. 3), and that the number of SNPs per scaffold correlated with the length of the scaffolds (R²=0.49) (Supplemental Fig. 2).
Identification of populations
PCAs were calculated for setA and setB. When including the Black Sea (setA), these individuals form a separate cluster and are highly divergent from samples from all other regions along the first principal component axis (PC1), which explains 3.7% of the total genomic variation (Fig. 2A). PC2 mainly separates the BALT individuals from ENA and WNA and explains 2.2% of total genomic variation. When excluding the Black Sea samples (setB), the sampling areas CA, ICE, NOS and SKA are not separated along PC1 and the Baltic Sea (BES and IBS) areas overlap partly (Fig. 2B). KAT is placed in-between the Baltic and the North Sea/North Atlantic sampling areas. This is congruent with its status as a transition zone, however, the amount of variation explained is low, i.e., PC1 and PC2 only explain 2.4% and 1.2% of the total genomic variation, respectively (Fig. 2B). The IBS partly overlaps with the BES, but some individuals also overlap with the KAT samples (Fig. 2B).
Principal Component Analyses: PC1 vs. PC2 of (A) SNP setA (incl. all sampling areas) and (B) setB (excluding the five Black Sea samples). Locality abbreviations refer to Fig. 1
Contrary to the PCA, the DAPC indicated stronger divergence among the samples. As a first step, we calculated the number of clusters found by the DAPC, which were three for setA and two for setB (Fig. 3A and B). For setA, we also see a clear separation of the Black Sea samples across DA1, which explains 89.7% of the variation between groups (Fig. 3C). DA2 separates BES from the others, but due to the strong signal from the Black Sea, the other sampling areas are not genetically distinguishable. When plotting DA3 against DA1, DA3 shows a separation of NOS from the other sampling areas (Fig. 3E). In the DAPC for setB, BES is distinct from the rest of the samples along DA1 (explaining 58% of between group variation), CA, ICE, and SKA overlap, slightly indicative of an isolation-by-distance (IBD) pattern, while NOS and IBS are set apart by DA2 (explaining 17% of the variation; Fig. 3D) and DA3 (explaining, 10% of the variation; Fig. 3F), respectively, and KAT is intermediate.
Discriminant analyses of principal components (DAPC). A, C and E include all samples (setA), B, D and F exclude the WBS samples (setB). Panel A + B are showing the assumed number and accordance to clusters. C and D are displaying the DA1 versus DA2. E and F are displaying the DA1 versus DA3. Locality abbreviations refer to Fig. 1
When using admixture and assuming a two-population scenario (k = 2, Fig. 4) for setB, one cluster (yellow) is formed by the Atlantic Sea Basins (CA, ICE, NOS, SKA and KAT), while the other (blue) is formed by the BALT region (BES and IBS), although IBS includes also some individuals assigned to the yellow cluster. When increasing k, assuming a three-population structure, more than half of the North Sea individuals are assigned to a third cluster (orange), to which – outside of the North Sea – only a single specimen in BES (Pool2-145) is assigned. Increasing k to 4 does not support any further geographic separation of the samples, as the “skyblue” cluster does not correlate with any specific sample characteristic (year, season, age or sex; Fig. 4, Supplement Table 1). Beside the one individual in BES, which displays high assignment to the orange cluster, a second individual sampled in NOS (Pool3-189) stands out with a high assignment to the blue cluster (Pool3-189). When excluding these two samples from the PCA analysis, ellipses of both NOS and BES become narrower and further apart (Supplemental Fig. 5). When including the Black Sea individuals (setA, Supplemental Fig. 4), they form a clearly distinguishable cluster on their own. For the North Atlantic and Baltic individuals, a similar cluster structure as with setB is revealed. The “orange” North Sea cluster only becomes apparent when increasing k to 5, however individuals are assigned to this cluster with high likelihood.
admixture plot based on SNP setB (145 samples, seven sampling areas). Each bar represents one individual, while the colour indicates genetic identity to the respective cluster. K2, k3 and k4 are shown. For k2 the clusters are coloured in blue and yellow. K3 adds orange and k4 adds skyblue. Locality abbreviations refer to Fig. 1
Using the software tableau, we projected the geographical occurrence of individual harbour porpoises assigned to the different clusters based on the admixture results (setB, k3) on to a map in accordance with the sampling coordinates of each specimen (Fig. 5). The yellow cluster, encompassing individuals from CA, ICE, NOS, SKA, KAT and IBS, occurs across all sea basins, except for the southern part of the Belt Sea, where the blue cluster dominates. The orange cluster is mainly restricted to the Eastern North Sea, except for one individual sampled in the Belt Sea. The Inner Baltic Sea exhibits individuals from both the yellow and the blue cluster, as well as admixed individuals. A similar pattern is shown in KAT, supporting its categorisation as a transition zone between North Atlantic and Belt Sea harbour porpoises.
Cluster assignment based on a 50% threshold for each sample, identified by admixture (k3) based on setB. Samples are plotted on a map according to their geographic coordinates. No specimens were admixed between the orange and the blue cluster. North Sea = Orange; Atlantic = yellow; Belt Sea = blue; Atlantic-North Sea admixed = red; Atlantic-Belt Sea admixed = green
In addition to the analysis of nuclear SNPs, we investigated the maternal population structure using the mitochondrial control region. A total of 387 bp was sequenced and 48 haplotypes could be identified among our 150 samples. 32 haplotypes are newly described here, while 16 matched the ones previously published (Lah et al. 2016; Wiemann et al. 2010). In the calculated haplotype network (Fig. 6), most haplotypes are separated by only one mutation step. The Black Sea samples show closely related haplotypes separated from the North Atlantic haplotypes. Black Sea haplotypes are accumulated on one side of the haplotype network, while Belt Sea and Inner Baltic Sea haplotypes are mostly found on the opposite side of the haplotype network. In-between, the placement of haplotypes follows a geographic pattern. Some haplotypes are shared among regions; however, haplotype frequencies differ drastically between the sampling areas. The WNA (CA and ICE) shows the most private haplotypes, while in both the BALT and ENA region overall fewer haplotypes are present. Some more abundant haplotypes differ in frequency among ENA and BALT, as PHO1 (56.7% in ENA and 27.3% in BALT), PHO4 (13% in ENA, absent in BALT) and PHO7 (6.7% in ENA and 48.5% in BALT; Supplemental Table 3).
Haplotype network of the mitochondrial control region. Each circle represents one haplotype, while the cipher represents the haplotype identification. Circle size indicates number of samples; colour is affiliation to sampling area. Bars indicate number of mutational steps between haplotypes. Locality abbreviations refer to Fig. 1
Genetic diversity and differentiation
Measures of genetic variability were assessed across all sampling areas considering SNP setA (Table 2). The results indicate only small differences in genetic diversity between the different sampling areas, except WBS, which shows the lowest values in observed heterozygosity and nucleotide diversity for both SNP and mtDNA (Table 2). When looking at the other sampling areas, the lowest observed heterozygosity is found in CA (HO=0.196 ± 0.004) and the highest in BES (HO=0.205 ± 0.007). Nominally, the observed heterozygosities were consistently slightly higher than the expected heterozygosities, but this deviation was not significant for any sampling area (Table 2). The nucleotide diversity was similar across sampling regions, with highest values detected in the BALT sampling areas, and lowest being observed in the WNA sampling areas. The ENA sampling areas showed an intermediate nucleotide diversity. For the mitochondrial control region (387 bp, 48HT), genetic diversity substantially differed across sampling areas. Here, CA and ICE had the highest nucleotide diversity (congruent with the highest number of haplotypes) while for the other sea basins the genetic diversity is considerably smaller. Both BALT sampling areas exhibited the lowest mtDNA nucleotide diversity.
Analyses of molecular variance (AMOVA) showed significant divergence among sampling areas for both mtDNA (FST = 0.310, p < 0.001) and SNPs (FST = 0.012, p < 0.001, Table 3). In the hierarchical AMOVA, the differentiation among regions for both marker sets were not significant (mtDNA: FCT = 0.294, p = 0.066; SNPs: FCT = 0.011, p = 0.063). Furthermore, sampling areas within regions were significantly divergent for SNPs (FSC = 0.001, p < 0.001), but not for the mtDNA (FSC = 0.023, p = 0.229; Table 3). Most genetic variation occurred for both marker sets within sampling areas (mtDNA = 69.0%, SNPs = 98.8%). Of the remaining variation, most was due to divergence among regions (mtDNA: 29.4%; SNPs: 1.12%).
Almost all pairwise FST estimates based on SNP setB were significant (α < 0.05* and α < 0.01**) after Bonferroni correction (except CA vs. ICE; Table 4). This result indicates genetic differentiation between the Atlantic sampling areas (WNA and ENA) and the Baltic sampling areas (BALT) with FST >0.01 for all comparisons. In contrast, FST estimates within the different regions (WNA, ENA and BALT) are considerably lower (e.g., CA vs. ICE: FST =0.000; NOS vs. SKA: FST =0.003, BES vs. IBS: FST = 0.006). The pairwise FST estimates based on only the mitochondrial control region were generally higher than the ones based on SNPs (Table 4). All pairwise comparisons with BES and almost all with CA (four out of five) were significant.
Detection of outlier regions and loci
The sliding window approach of the FST allows identifying genomic regions where SNPs exhibit high FST values (Fig. 7). The average FST for all windows across all scaffolds is 0.28 (blue line in Fig. 7). For the 34 largest scaffolds, we plotted FST values for all 500 kb windows, along with scaffold-specific 95% confidence intervals. The rationale here is to identify those scaffolds with generally lower FST values (e.g. ScshVNz_10623, ScshVNz_7191, ScshVNz_7648, Fig. 7) and respectively generally higher FST values (e.g. ScshVNz_11228, Fig. 7), as compared to the genome-wide mean FST (blue line). Within scaffolds, we can further identify regions where the calculated FST of one or more sliding windows is outside the 95% confidence interval of the respective scaffold, depicting genomic regions of low and respectively high divergence (e.g. in ScshVNz_10729, ScshVNz_11943, ScshVNz_12150, Fig. 7).
Single plots for each of the 34 largest scaffolds, showing the FST, calculated as sliding window of 500 kb with popgen, based on SNP setB (Black Sea excluded). X-values of red dots depict the position of the sliding windows on the respective scaffold, blue line indicates the average FST over all 34 scaffolds, the black dashed lines indicate the upper and lower bounds of the confidence limit for each separate scaffold individually. mbp = million base pairs
With population genomic data at hand, we can also investigate if SNPs are indicative for the different found clusters or sampling areas, and if so, investigate if they occur in coding regions or are under selection. We could identify six SNPs with statistically significant patterns of divergent genetic differentiation (having an α-value of greater than ten or a FST value greater then 0.05, respectively; Supplemental Fig. 6). The allele frequencies of these SNPs are congruent with isolation-by-distance across the different sampling areas throughout the North Atlantic (Table 5, Supplemental Table 2). This becomes even more apparent when looking at the three main ocean basins/regions (Fig. 8). Although none of the six outliers are positioned within coding regions of genes in the nuclear genome or their flanking regions (as far as predicted by the draft annotation), the positive alpha indicates that they may all be influenced by diversifying selection with regard to sampling regions.
Discussion
Population differentiation in the North Atlantic, relative to the divergence from the Black Sea
The population structure of the harbour porpoise within the North Atlantic is under ongoing investigation (NAMMCO 2019). Long-distance movements are known to occur (Jefferson 2014; Nielsen et al. 2019), which can obscure subtle, genetic differentiation between oceanic regions (Leslie and Morin 2016). Although the North Atlantic is predominantly an open water body with few to no physical barriers, potentially allowing for harbour porpoises of different geographic origin to intermix freely and to successfully mate with each other, harbour porpoise are known to be shallow water cetaceans avoiding water bodies deeper than 300 m. Thus, deep waters, e.g. the Icelandic basin or the Norwegian Sea, may act as natural barriers, forcing migration to follow the Greenland-Scotland ridge. These limitations could have supported differentiation in harbour porpoise, whose range within the North Atlantic has been subdivided into multiple distinct management areas that have been delineated based on genetics, morphometry, as well as pollutant profiles and telemetry data (Fontaine et al. 2007, 2014; Lah et al. 2016; NAMMCO 2019; Nielsen et al. 2018; Rosel et al. 1999a; Wiemann et al. 2010; Celemin et al. 2023). Within the North Atlantic, these inferred separations are suggested to be demographically independent, with finer subdivisions in the East North Atlantic, including North and Baltic Seas. The NAMMCO assessment units are currently supported by the identification of isolation-by-distance across the North Atlantic up into the Baltic Sea (Fontaine et al. 2007; Lah et al. 2016).
Here we used genome-wide distributed SNPs to unravel potential population structure within the entire North Atlantic distribution range of the species. We find very little population structure between Canada and Iceland, which - together with findings of Celemin et al. 2023; Fontaine et al. 2017; Olsen et al. 2022; and Quintela et al. 2020 - suggests gene flow across most of the North Atlantic from Canada to Norway. However, other studies have revealed differentiation in the Western North Atlantic involving areas not sampled in our study, i.e., between the US Atlantic coast and Canada, and Western Greenland harbours a porpoise population genetically distinct from those of Canada and Iceland (NAMMCO 2019; Nielsen et al. 2018; Olsen et al. 2022; Tolley et al. 2001).
Our mtDNA data and a previous mtDNA study (Rosel et al. 1999) hint at differentiation between East and West North Atlantic, as very few haplotypes are shared between the Western North Atlantic and the Eastern North Atlantic/Baltic Sea regions (PHO1, 4, 19, 20; cf. Figure 6). While in WNA a higher number of haplotypes occurs, all in frequencies below 10%, we find high haplotype frequencies for few haplotypes (PHO1 and PHO4) in ENA. This representation of more maternal lines in WNA could be indicative of a higher long-term effective population size (Ne), while ENA/BALT exhibits a star-like mtDNA phylogeny, pointing towards recent bottlenecks/colonization events with subsequent expansions (Fig. 6). Such differences in long-term Ne are also indicated by the about three times larger mtDNA nucleotide diversity in WNA, when compared to the other regions.
Comparing nuclear (SNP) with mtDNA data we observed a contrasting pattern: SNP divergence is most pronounced between the Baltic region and the open ocean regions (North Sea, North Atlantic), with only subtle isolation-by-distance across the entire North Atlantic. It may hence reflect adaptive rather than mere geographic processes. Conversely, mtDNA is most differentiated between West and East North Atlantic, indicative of higher philopatry in females, as has been postulated repeatedly (e.g., Wiemann et al. 2010). Ocean-wide connectivity may therefore be mostly driven by occasional male dispersal which does not contribute to the observed mtDNA pattern, unless the migrating individual itself is sampled (Tiedemann et al. 2000).
Although there is apparently only little differentiation at SNP markers across the North Atlantic, marginal and atypical environments could lead to genetic differentiation, such as shown around the UK (Fontaine et al. 2017). The region encompasses strikingly different marine habitats from oceanic to the rather shallow North Sea, and a transition into the more brackish habitats of the Baltic Sea, so the detection of cryptic population structure would not be surprising. Here, we detect subtle differences between the Eastern North Sea (NOS) and other North Atlantic porpoises. The Skagerrak porpoises (SKA) are closer to Icelandic ones (ICE) than to the close-by North Sea (NOS), both in the DAPC (Fig. 3D and F) and regarding pairwise FSTs (Table 4). Moreover, some NOS specimens form a separate genetic cluster in the admixture plot (Fig. 4). This assignment becomes even more prominent when the outgroup Black Sea samples are included in the analysis (Supplemental Fig. 4). The North Sea samples responsible for this signal of differentiation are mainly occurring in the German Wadden Sea, around the Isle of Sylt (Fig. 5). This is in accordance with previous investigations, which identified a distinct breeding ground around the Isle of Sylt (Diederichs et al. 2010; Sonntag et al. 1999; Unger et al. 2022). A more complete sampling of the North Sea harbour porpoises is needed to assess the robustness as well as the geographic distribution of a separate North Sea cluster.
Additional local population structure is detected for the Baltic region (BALT; here defined as BES and IBS), which is separated from the North Sea/North Atlantic. ADMIXTURE distinguishes here between a Baltic and a North Atlantic cluster (“blue” and “yellow” in Fig. 4, respectively), which are also identified in the DAPC (Fig. 3B). This separation is further supported by high FST values (Table 4). The Kattegat (KAT) sampling area represents a transition zone between these two regions. Also, significant differences in the frequencies of particular mitochondrial haplotypes support this assignment, i.e. PHO4 (19% in NOS vs. 0% in BALT and 5% in WNA) and PHO7 (BES 50%, IBS 41.7% vs. NOS 9.5%, CA, ICE, SKA = 0%). The disproportional geographic distribution of these two haplotypes has been previously recognized and correlates with divergence at nuclear loci, both SNPs and microsatellites (Lah et al. 2016; Wiemann et al. 2010).
A further subdivision within the Baltic (BALT) region based on differences in genetics, morphology, and behavior has been repeatedly hypothesized, such that porpoises from BES and IBS would belong to two distinct populations (Celemin et al. 2023; Galatius et al. 2012; Lah et al. 2016; NAMMCO 2019; Wiemann et al. 2010). This could be caused by a separation of these two groups during mating season and calving (Carlén et al. 2018; Huggenberger et al. 2002). Our data identify some genetic differences among these two Baltic sampling areas with strong support for a Belt Sea population. However, the inner Baltic specimens were not unambiguously identified as a discrete population, as these differences may also be reconciled by a mixture of migrating specimens of both Belt Sea and North Atlantic/Skagerrak origin (cf. Figure 5). Of note in this context, we defined IBS according to Wiemann et al. (2010), i.e., east of 13.5°E longitude and our 12 IBS samples originate from the westernmost part of this area, south of Sweden. Recent studies suggest the distinct IBS population to occur further to the east (Carlén et al. 2018), such that only our three easternmost individuals originate from the distribution range of that putative population (Fig. 5), too few to reach any conclusion about the status of this population. Genomic studies including samples from further East (Poland, East of Gotland) assigned some Inner Baltic specimens to a separate cluster (Celemin et al. 2023; Lah et al. 2016).
Haplotype frequencies in both the ENA (PHO4 + PHO1) and BALT (PHO7 + PHO1) regions could indicate a more recent expansion, as we see a star like pattern in the haplotype network (Fig. 6) with common widespread ancestral haplotypes and closely related locally unique, rare haplotypes (Rosel et al. 1999; Wiemann et al. 2010). This would be consistent with the history of the Baltic Sea that was only recently (i.e., several thousand years ago) accessible to harbour porpoises. Previous studies have detected a similar pattern in further ENA regions, i.e., Norway (Tolley and Rosel 2006) and around the UK (Rosel et al. 1999; Walton 1997).
By identifying genetic clusters without a priori geographic assignment, our data allows the detection of migrating individuals. The harbour porpoise is known to seasonally migrate in some regions in response to the formation of sea-ice during winter, such as in the Labrador Sea between Canada and Greenland (Olsen et al. 2022), but also in the Northern part of the proper Baltic Sea (Andersen et al. 2001; Benjamins et al. 2007; Dähne et al. 2017; Nielsen et al. 2018; Rosel et al. 1999a). Seasonal migrations may also correlate with breeding behavior, as mating takes place mostly in the summer months (May to October), and animals migrate between breeding and winter feeding grounds in some areas, e.g. in the Baltic Sea (Carlén et al. 2018; Jefferson 2014; Sveegaard et al. 2015). Females tend to come back to regions where they were born for mating and calving, which in consequence limits the gene flow between populations (Andersen et al. 2001; Huggenberger et al. 2002; Kesselring et al. 2017). Gene flow could be maintained by single dispersing individuals, which may be primarily males (Huggenberger et al. 2002; NAMMCO 2019; Wiemann et al. 2010). In our study, we find two individuals, which show a strong genetic association with a cluster from a geographic region where they were not sampled from, indicating potential migrants. Within the NOS, one sample (Pool3-189, comprising a male sampled in May) shows a strong cluster affiliation to the Belt Sea (blue, Figs. 4 and 5) and has the mitochondrial PHO7, indicative for the BALT region. Therefore, we assume it to be a Belt Sea individual, which migrated into the North Sea. We found one female individual sampled in the Belt Sea (Pool2-145), which seemed to have migrated into the opposite direction. This sample has a high assignment probability to the orange cluster (NOS, Figs. 4 and 5) and carries the PHO5, a rather rare haplotype known to occur both in North and Belt Sea (Tiedemann et al. 1996; Wiemann et al. 2010). When excluding these two putative migrant samples from the dataset, the cluster assignment in the PCA is improved (Supplemental Fig. 5).
Population differentiation among our defined regions (WNA, ENA, BALT) is also supported by the six outlier SNP loci, which clearly distinguish them (Fig. 8). SNPs 1, 3, 4 and 6 further differentiate between WNA and ENA (Fig. 8). Additionally, these SNPs support the previously shown isolation-by-distance across the North Atlantic (Fontaine et al. 2007; Galatius et al. 2012; Lah et al. 2016; Tolley and Rosel 2006). Although the population structure pattern found in this study by SNP and mtDNA data suggest potential local adaptation, the outlier SNPs do not lie in coding regions of the genome but could be linked to loci under selection. Hence, further studies using full nuclear genome analyses to identify candidate genes for local adaptation should be performed.
Implication for conservation
With our mtDNA data we can detect a subtle differentiation between WNA and ENA, but this pattern was not observed in the SNP data. The substantially higher genetic variation in terms of nucleotide diversity of mtDNA in WNA as compared to the other regions (cf. Table 2) suggests a higher effective population size there. This is in line with high abundance estimates derived from surveys, as well as with the genetic cohesion over a wide geographical range, encompassing Canadian to Icelandic waters. Intriguingly, more subtle population structure exists on the Eastern side of the Atlantic, as we detected differentiation between the respective sampling areas within ENA and BALT with a transition zone in the Kattegat in both mtDNA and SNPs. The different haplotype frequencies and high occurrence of specific indicative haplotypes, especially in NOS, BES and IBS, significantly deviate from a random distribution and therefore support demographic independence of the populations inhabiting these different sampling areas. Intriguingly, we found some support for a separate genetic cluster in the German North Sea, coinciding with the observation of a distinct breeding ground around the Isle of Sylt (Gilles et al. 2009; Siebert et al. 2006; Sonntag et al. 1999). Although the full geographic range occupied by porpoises belonging to this inferred cluster could not be revealed here, specific protection measures for this area may be warranted, not least as this area is subject to increasing construction of offshore wind parks, which increase the disturbance for the local marine fauna (Aarts et al. 2016; Booth 2020; Dähne et al. 2017; Peschko et al. 2016; Schaffeld et al. 2020). Although abundances in the North Atlantic shelf distribution area are quite high, the genetic differentiation shown here, as well as the identified morphological and behavioral differences previously observed may warrant consideration of more regional management units for which specific abundance estimates would be desirable. This was done for the BALT region, where the abundance estimation is separated for BES (~ 14,400) and IBS (~ 500 animals; Benke et al. 2014; Gilles et al. 2023; Hammond et al. 2018; SAMBAH 2016). It was shown, that the Inner Baltic (IBS) porpoises migrate between feeding and breeding grounds, and that the breeding time distribution does not overlap between Inner Baltic and Belt Sea (Carlén et al. 2018). The exact geographical split among these populations remains elusive, but is considered more eastwards than previously thought (Carlén et al. 2018). Our data generally supports the assessment areas of NAMMCO (NAMMCO 2019) and warrants consideration of the southern NOS, BES and IBS as separate populations/management units, with a recommendation to include further samples from neighbouring areas in future studies.
Genome-wide population assessments like ours can be utilized to establish SNP assays specifically tailored to distinguish between individuals assigned to the different identified genetic clusters. When designing such a SNP assay, inferred genetic clusters need to be related to geographic populations (here, North Atlantic, yellow cluster; Belt Sea, blue cluster; and southern North Sea, orange cluster) and potential migrants (i.e., specimens with a genetic cluster assignment different from the inferred population of their sampling area) should be excluded. Sample size permitting, one may restrict this inference to specimens originating from the reproductive season, as these have a higher likelihood to belong to the local population (e.g., Wiemann et al. 2010). Then, the focus should be on private/population specific SNPs, i.e. such as our detected outlier SNPs. These outlier loci can already be considered as a candidate panel to estimate differential assignment likelihoods to the respective populations. Additionally, we could detect regions on different scaffolds, where SNPs with high or low FST accumulated. Therefore, we identified genomic regions, which are potentially more indicative for population separation than others (Supplemental Fig. 3). SNPs from these regions would also be candidates for the design of such a panel. As our data set was used as training data to inform that panel, it needs to be validated by an independent test data set of known geographic affiliation. Once validated, a SNP assay could also identify migrants between the different seas and potentially provide genetic profiles from stranded decomposed animals that may not be easily genotyped using other methods. This would enable to infer the impact of bycatch and other mortality incidences to specific potentially threatened geographic populations, thereby becoming a valuable tool in conservation and management of porpoises.
Data Availability
Sequence information for all SNPs is provided in supplemental table 2. The underlying sequence data (raw reads) have been submitted to Genbank. Reference mtDNA sequences (Pho1-Pho9) are available at Genbank (accession no. Y13872.1-Y13880.1).
References
Aarefjord H, Björge A, Kinze CC, Lindstedt I (1995) Diet of the harbour porpoise (Phocoena phocoena) in scandinavian waters. Rep Int Whal Commn Spec 16:211–222
Aarts G, Von Benda-Beckmann AM, Lucke K, Sertlek HÖ, Van Bemmelen R, Geelhoed SCV, …, Kirkwood R (2016) Harbour porpoise movement strategy affects cumulative number of animals acoustically exposed to underwater Explosions. Mar Ecol Prog Ser 557:261–275. https://doi.org/10.3354/meps11829
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19(9):1655–1664. https://doi.org/10.1101/gr.094052.109
Andersen LW, Ruzzante DE, Walton M, Berggren P, Bjørge A, Lockyer C (2001) Conservation genetics of harbour porpoises, Phocoena phocoena, in eastern and central North Atlantic. Conserv Genet, (2), 309–324
ASCOBANS (2002) Recovery Plan for Baltic Harbour porpoises (Jastarnia Plan). ASCOBANS Rep 83(July):5–9
ASCOBANS (2021) Special species Sessions Harbour Porpoise Baltic Proper & Iberian Populations. Draft proposal for the inclusion of the Iberian Harbour Porpoise on the appendices of CMS. ASCOBANS Rep 44(October):8–12
Attard CRM, Beheregaray LB, Sandoval-Castillo J, Jenner KCS, Gill PC, Jenner MNM, …, Möller LM (2018) From conservation genetics to conservation genomics: a genome-wide assessment of blue whales (Balaenoptera musculus) in Australian feeding aggregations. Royal Soc Open Sci 5(1). https://doi.org/10.1098/rsos.170925
Autenrieth M, Hartmann S, Lah L, Roos A, Dennis AB, Tiedemann R (2018) High-quality whole-genome sequence of an abundant holarctic odontocete, the harbour porpoise (Phocoena phocoena). Mol Ecol Resour 18(6):1469–1481. https://doi.org/10.1111/1755-0998.12932
Baker CS, Steel D, Calambokidis J, Falcone E, González-peral U, Barlow J, …, Yamaguchi M (2013) Strong maternal fidelity and natal philopatry shape genetic structure in North Pacific humpback whales. Mar Ecol Prog Ser 494:291–306. https://doi.org/10.3354/meps10508
Benjamins S, Lawson J, Stenson G (2007) Recent harbour porpoise bycatch in gillnet fisheries in Newfoundland and Labrador, Canada. J Cetacean Res Manage 9(3):189–199
Benke H, Bräger S, Dähne M, Gallus A, Hansen S, Honnef CG, …, Verfuß UK (2014) Baltic Sea harbour porpoise populations: Status and conservation needs derived from recent survey results. Mar Ecol Prog Ser 495:275–290. https://doi.org/10.3354/meps10538
Booth CG (2020) Food for thought: Harbor porpoise foraging behavior and diet inform vulnerability to disturbance. Mar Mamm Sci 36(1):195–208. https://doi.org/10.1111/mms.12632
Cammen KM, Andrews KR, Carroll EL, Foote AD, Humble E, Khudyakov JI, …, Van Cise AM (2016) Genomic methods take the plunge: recent advances in high-throughput sequencing of marine mammals. J Hered 107(6):481–495. https://doi.org/10.1093/jhered/esw044
Carlén I, Thomas L, Carlström J, Amundin M, Teilmann J, Tregenza N, …, Acevedo-Gutiérrez A (2018) Basin-scale distribution of harbour porpoises in the Baltic Sea provides basis for effective conservation actions. Biological Conservation, 226(May 2017), 42–53. https://doi.org/10.1016/j.biocon.2018.06.031
Carlström J, Carlén I, Dähne M, Koschinski S, Owen K, Sveegaard S, …, Hammond PS (2023) Phocoena phocoena (Linnaeus, 1758) (Baltic Proper Subpopulation) Harbour porpoise Red List Assessment. IUCN Red List of Threatened Species. Version 2014.3
Carroll EL, Reyes C, Gaggiotti OE, Olsen MT, Maaholm DJ, Rosso M, Davison N (2016) Pilot study to assess the utility of ddRAD sequencing in identifying species-specific and shared SNPs among Blainville’s (Mesoplodon densirostris) and Cuvier’s (Ziphius cavirostris) beaked whales. Int Whal Comm. https://doi.org/10.13140/RG.2.1.2286.5527
Celemin E, Autenrieth M, Roos A, Pawliczka I, Lindstr U, Benke H, …, Tiedemann R (2023) Evolutionary history and seascape genomics of Harbour porpoises (Phocoena phocoena) across environmental gradients in the North Atlantic and adjacent waters. Molecular Ecology Resources, in press
Çilingir FG, Hansen D, Bunbury N, Postma E, Baxter R, Turnbull L, …, Grossen C (2022) Low-coverage reduced representation sequencing reveals subtle within-island genetic structure in Aldabra giant tortoises. Ecol Evol 12(3):1–13. https://doi.org/10.1002/ece3.8739
Cornish A, Guda C (2015) A Comparison of Variant Calling Pipelines Using Genome in a Bottle as a Reference. BioMed Research International, 2015. https://doi.org/10.1155/2015/456479
Dähne M, Tougaard J, Carstensen J, Rose A, Nabe-Nielsen J (2017) Bubble curtains attenuate noise from offshore wind farm construction and reduce temporary habitat loss for harbour porpoises. Mar Ecol Prog Ser 580:221–237. https://doi.org/10.3354/meps12257
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12(7):499–510. https://doi.org/10.1038/nrg3012
de Greef E, Einfeldt AL, Miller PJO, Ferguson SH, Garroway CJ, Lefort KJ, …, Feyrer LJ (2022) Genomics reveal population structure, evolutionary history, and signatures of selection in the northern bottlenose whale, Hyperoodon ampullatus. Mol Ecol 31(19):4919–4931. https://doi.org/10.1111/mec.16643
Diederichs A, Brandt MJ, Nehls G (2010) Does sand extraction near Sylt affect harbour porpoises ? Wadden Sea Ecosystem 26(26):199–203
Dungan SZ, Wang JY, Araújo CC, Yang SC, White BN (2016) Social structure in a critically endangered Indo-Pacific humpback dolphin (Sousa chinensis) population. Aquat Conservation: Mar Freshw Ecosyst 26(3):517–529. https://doi.org/10.1002/aqc.2562
Durban JW, Elston DA, Ellifrit DK, Dickson E, Hammond PS, Thompson PM (2005) Multisite mark-recapture for cetaceans: Population estimates with bayesian model averaging. Mar Mamm Sci 21(1):80–92. https://doi.org/10.1111/j.1748-7692.2005.tb01209.x
Dyndo M, Wiśniewska DM, Rojano-Doñate L, Madsen PT (2015) Harbour porpoises react to low levels of high frequency vessel noise. Sci Rep 5:11083. https://doi.org/10.1038/srep11083
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3):564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Farmer NA, Baker K, Zeddies DG, Denes SL, Noren DP, Garrison LP, …, Zykov M (2018) Population consequences of disturbance by offshore oil and gas activity for endangered sperm whales (Physeter macrocephalus). Biol Conserv 227(September):189–204. https://doi.org/10.1016/j.biocon.2018.09.006
Foll M (2012) BayeScan v2.1. Ecology 20(11):1450–1462. https://doi.org/10.1111/2041-210X.12418
Foll M, Gaggiotti O (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a bayesian perspective. Genetics 180(2):977–993. https://doi.org/10.1534/genetics.108.092221
Fontaine MC, Baird SJ, Piry S, Ray N, Tolley KA, Duke S, …, Michaux JR (2007) Rise of oceanographic barriers in continuous populations of a cetacean: the genetic structure of harbour porpoises in Old World waters. BMC Biol 5(1):30. https://doi.org/10.1186/1741-7007-5-30
Fontaine MC, Tolley KA, Michaux JR, Birkun A, Ferreira M, Jauniaux T, …, Baird SJE (2010) Genetic and historic evidence for climate-driven population fragmentation in a top cetacean predator: the harbour porpoises in European water. Proceedings. Biological Sciences / The Royal Society, 277, 2829–2837. https://doi.org/10.1098/rspb.2010.0412
Fontaine MC, Roland K, Calves I, Austerlitz F, Palstra FP, Tolley KA, …, Aguilar A (2014) Postglacial climate changes and rise of three ecotypes of harbour porpoises, Phocoena phocoena, in western Palearctic waters. Mol Ecol 23(13):3306–3321. https://doi.org/10.1111/mec.12817
Fontaine MC, Thatcher O, Ray N, Piry S, Brownlow A, Davison NJ, …, Goodman SJ (2017) Mixing of porpoise ecotypes in southwestern UK waters revealed by genetic profiling. Royal Soc Open Sci 4(3):1–14. https://doi.org/10.1098/rsos.160992
Galatius A, Kinze CC, Teilmann J (2012) Population structure of harbour porpoises in the Baltic region: evidence of separation based on geometric morphometric comparisons. J Mar Biol Association United Kingd 92(8):1669–1676. https://doi.org/10.1017/S0025315412000513
Gallego-García N, Caballero S, Shaffer HB (2021) Are genomic updates of well-studied Species Worth the investment for Conservation? A case study of the critically endangered Magdalena River Turtle. J Hered 112(7):575–589. https://doi.org/10.1093/jhered/esab063
Gao H, Kang H, Zhang Y, Wang J, Lin W, Zhang P, …, Li S (2023) Chromosome-Level Genome Assembly of the rough-toothed Dolphin (Steno bredanensis). J Mar Sci Eng 11(2). https://doi.org/10.3390/jmse11020418
Gilles A, Scheidat M, Siebert U (2009) Seasonal distribution of harbour porpoises and possible interference of offshore wind farms in the German North Sea. Marine Ecology Progress Series, 383(June 2008), 295–307. https://doi.org/10.3354/meps08020
Gilles A, Gunnlaugsson T, Mikkelsen B, Pike DG, Víkingsson GA (2020) Summer Abundance of Harbour Porpoises (Phocoena phocoena) in the Coastal Waters of Iceland and the Faroe Islands. NAMMCO Scientific Publications, 11. https://doi.org/10.7557/3.4939
Gilles A, Authier M, Ramirez-Martinez NC, Araújo H, Blanchard A, Carlström J, Eira C, Dorémus G, Fernández-Maldonado C, Geelhoed SCV, Kyhn L, Laran S, Nachtsheim D, Panigada S, Pigeault R, Sequeira M, Sveegaard S, Taylor NL, Owen K, Saavedra C, Vázquez-Bonales JA, Unger B, Hammond PS (2023) Estimates of cetacean abundance in European Atlantic waters in summer 2022 from the SCANS-IV aerial and shipboard surveys. Final report published 29 September 2023. 64 pp. https://tinyurl.com/3ynt6swa
Goudet J (2005) HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186. https://doi.org/10.1111/j.1471-8278
Guzman HM, Gomez CG, Guevara CA, Kleivane L (2013) Potential vessel collisions with Southern Hemisphere humpback whales wintering off Pacific Panama. Mar Mamm Sci 29(4):629–642. https://doi.org/10.1111/j.1748-7692.2012.00605.x
Hammond PS, Macleod K, Berggren P, Borchers DL, Burt L, Canadas A, …, Vazquez JA (2013) Cetacean abundance and distribution in European Atlantic shelf waters to inform conservation and management. Biol Conserv 164:107–122. https://doi.org/10.1016/j.biocon.2013.04.010
Hammond PS, Lacey C, Gilles A, Viquerat S, Börjesson P, Herr H, …, Olen N (2018) Estimates of cetacean abundance in European Atlantic waters in summer 2016 from the SCANS-III aerial and shipboard surveys. Wageningen Mar Res, (May), 1–40
Hammond PS, Bearzi G, Bjørge A, Forney KA, Karczmarski L, Kasuya T, …, Wilson B (2020) Phocoena phocoena (Baltic Sea subpopulation). (errata version published in 2016). Retrieved from The IUCN Red List of Threatened Species website: http://www.iucnredlist.org/details/17031/0
Härkönen T, Galatius A, Bräeger S, Karlsson O, Ahola M (2013) Population growth rate, abundance and distribution of marine mammals. HELCOM Core Indicator of Biodiversity Population 1–34. https://doi.org/10.1128/AAC.03728-14
Holt MM, Hanson MB, Giles DA, Emmons CK, Hogan JT (2017) Noise levels received by endangered killer whales Orcinus orca before and after implementation of vessel regulations. Endanger Species Res 34:15–26. https://doi.org/10.3354/esr00841
Huggenberger S, Benke H, Kinze CC (2002) Geographical variation in harbour porpoise (Phocoena phocoena) skulls: support for a separate non-migratory population in the Baltic proper. Ophelia 56(1):1–12. https://doi.org/10.1080/00785236.2002.10409484
IJsseldijk LL, ten Doeschate MTI, Brownlow AC, Davison NJ, Deaville R, Galatius A, …, Heesterbeek H (2020) Spatiotemporal mortality and demographic trends in a small cetacean: strandings to inform conservation management. Biol Conserv March108733. https://doi.org/10.1016/j.biocon.2020.108733
Jefferson TA (2014) Phocoenidae (Porpoises). Wilson, D.E. Mittermeier, R.A., Handbook of the mammals of the World - volume 4. Lynx Edicions, pp 528–548
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24(11):1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Kesselring T, Viquerat S, Brehm R, Siebert U (2017) Coming of age: - do female harbour porpoises (Phocoena phocoena) from the North Sea and Baltic Sea have sufficient time to reproduce in a human influenced environment? PLoS ONE 12(10):e0186951. https://doi.org/10.1371/journal.pone.0186951
Korneliussen TS, Albrechtsen A, Nielsen R (2014) ANGSD: analysis of next generation sequencing data. BMC Bioinformatics 15(1):1–13. https://doi.org/10.1186/s12859-014-0356-4
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Kumaran M, Subramanian U, Devarajan B (2019) Performance assessment of variant calling pipelines using human whole exome sequencing and simulated data. BMC Bioinformatics 20(1):1–11. https://doi.org/10.1186/s12859-019-2928-9
Lah L, Trense D, Benke H, Berggren P, Gunnlaugsson Þ, Lockyer C, …, Tiedemann R (2016) Spatially explicit analysis of genome-wide SNPs detects subtle Population structure in a Mobile Marine Mammal, the Harbor Porpoise. PLoS ONE 11(10):1–23. https://doi.org/10.1371/journal.pone.0162792
Leigh JW, Bryant D (2015) POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6(9):1110–1116. https://doi.org/10.1111/2041-210X.12410
Leslie MS, Morin PA (2016) Using genome-wide SNPs to detect structure in high-diversity and low-divergence populations of severely impacted eastern tropical pacific spinner (Stenella longirostris) and pantropical spotted dolphins (S. attenuata). Front Mar Sci 3(DEC):1–12. https://doi.org/10.3389/fmars.2016.00253
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26(5):589–595. https://doi.org/10.1093/bioinformatics/btp698
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, …, Durbin R (2009) The sequence Alignment/Map format and SAMtools. Bioinformatics 25(16):2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Liu N, Chen L, Wang S, Oh C, Zhao H (2005) Comparison of single-nucleotide polymorphisms and microsatellites in inference of population structure. BMC Genetics, 6(SUPPL.1), 1–5. https://doi.org/10.1186/1471-2156-6-S1-S26
Lonergan M, Duck CD, Thompson D, Moss S, McConnell B (2011) British grey seal (Halichoerus grypus) abundance in 2008: an assessment based on aerial counts and satellite telemetry. ICES J Mar Sci 68:2201–2209. https://doi.org/10.1093/icesjms/fsr161
Luca M, Andrew W, Emer R, Patricia R, Andrew R, Jamie C, Tom C (2009) Population structure of short-beaked common dolphins (Delphinus delphis) in the North Atlantic Ocean as revealed by mitochondrial and nuclear genetic markers. Mar Biol 156(5):821–834. https://doi.org/10.1007/s00227-008-1120-y
Maruki T, Lynch M (2017) Genotype calling from population-genomic sequencing data. G3: Genes, Genomes, Genetics, 7(5), 1393–1404. https://doi.org/10.1534/g3.117.039008
McCauley DJ, Pinsky ML, Palumbi SR, Estes JA, Joyce FH, Warner RR (2015) Marine defaunation: animal loss in the global ocean. Science 347(6219). https://doi.org/10.1126/science.1255641
McMahon BJ, Teeling EC, Höglund J (2014) How and why should we implement genomics into conservation? Evol Appl 7(9):999–1007. https://doi.org/10.1111/eva.12193
Mielczarek M, Szyda J (2016) Review of alignment and SNP calling algorithms for next-generation sequencing data. J Appl Genet 57(1):71–79. https://doi.org/10.1007/s13353-015-0292-7
Morin PA, Martien KK, Taylor BL (2009) Assessing statistical power of SNPs for population structure and conservation studies. Mol Ecol Resour 9(1):66–73. https://doi.org/10.1111/j.1755-0998.2008.02392.x
NAMMCO NAMMC (2019) and the N. I. of M. R. Report of the Joint IMR/NAMMCO International Workshop on the Status of Harbour Porpoises in the North Atlantic. Tromsø, Norway
Neely BA, Ellisor DL, Davis WC (2018) Proteomics as a metrological tool to evaluate genome annotation accuracy following de novo genome assembly: a case study using the Atlantic bottlenose dolphin (Tursiops truncatus). BioRxiv, 254250. https://doi.org/10.1101/254250
Nielsen NH, Teilmann J, Sveegaard S, Hansen RG, Sinding MHS, Dietz R, Heide-Jørgensen MP (2018) Oceanic movements, site fidelity and deep diving in harbour porpoises from Greenland show limited similarities to animals from the North Sea. Mar Ecol Prog Ser 597(November):259–272. https://doi.org/10.3354/meps12588
Nielsen NH, Teilmann J, Heide-Jørgensen MP (2019) Indications of mesopelagic foraging by a small odontocete. Mar Biol 166(6):1–8. https://doi.org/10.1007/s00227-019-3525-1
Nuuttila HK, Courtene-Jones W, Baulch S, Simon M, Evans PGH (2017) Don’t forget the porpoise: acoustic monitoring reveals fine scale temporal variation between bottlenose dolphin and harbour porpoise in Cardigan Bay SAC. Mar Biol 164(3):1–16. https://doi.org/10.1007/s00227-017-3081-5
Olsen MT, Nielsen NH, Biard V, Teilmann J, Ngô MC, Víkingsson G, …, Heide-Jørgensen MP (2022) Genetic and behavioural data confirm the existence of a distinct harbour porpoise ecotype in West Greenland. Ecological Genetics and Genomics, 22(November 2021), 100108. https://doi.org/10.1016/j.egg.2021.100108
Omeyer LCM, Doherty PD, Dolman S, Enever R, Reese A, Tregenza N, …, Godley BJ (2020) Assessing the effects of Banana pingers as a Bycatch Mitigation device for Harbour porpoises (Phocoena phocoena). Front Mar Sci 7(May):1–10. https://doi.org/10.3389/fmars.2020.00285
Paasche Ø, Österblom H, Neuenfeldt S, Bonsdorff E, Brander K, Conley DJ, …, Stenseth NC (2015) Connecting the seas of Norden. Nat Clim Change 5(2):89–92. https://doi.org/10.1038/nclimate2471
Paradis E (2010) Pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26(3):419–420. https://doi.org/10.1093/bioinformatics/btp696
Pembleton LW, Cogan NOI, Forster JW (2013) StAMPP: an R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol Ecol Resour 13(5):946–952. https://doi.org/10.1111/1755-0998.12129
Peschko V, Ronnenberg K, Siebert U, Gilles A (2016) Trends of harbour porpoise (Phocoena phocoena) density in the southern North Sea. Ecological Indicators, 60(July 2005), 174–183. https://doi.org/10.1016/j.ecolind.2015.06.030
Pierce GJ, Santos MB, Cerviño S (2007) Assessing sources of variation underlying estimates of cetacean diet composition: a simulation study on analysis of harbour porpoise diet in Scottish (UK) waters. J Mar Biol Association United Kingd 87(1):213–221. https://doi.org/10.1017/S0025315407055348
Pike D, Gunnlaugsson T, Sigurjonsson J, Vikingsson G (2020) Distribution and abundance of cetaceans in Icelandic Waters over 30 years of aerial surveys. NAMMCO Sci Publications 11:1–22. https://doi.org/10.7557/3.4805
Pritchard JK, Stephens M, Donnelly P (2000) Inference of Population structure using multilocus genotype data. Genet Soc Am 155:945–959
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, …, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3):559–575. https://doi.org/10.1086/519795
Quintela M, Besnier F, Seliussen B, Glover KA, Lindstrøm U (2020) Population structure of bycaught harbour porpoise (Phocoena phocoena) in Norway. Mar Biol Res 16(3):141–147. https://doi.org/10.1080/17451000.2020.1729992
Reeves IM, Totterdell JA, Barceló A, Sandoval-Castillo J, Batley KC, Stockin KA, …, Möller LM (2022) Population genomic structure of killer whales (Orcinus orca) in Australian and New Zealand waters. Mar Mamm Sci 38(1):151–174. https://doi.org/10.1111/mms.12851
Rochette NC, Colón AGR, Catchen JM, Rivera-Colón AG, Catchen JM (2019) Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol Ecol 28(21):4737–4754. https://doi.org/10.1111/mec.15253
Rolland RM, Parks SE, Hunt KE, Castellote M, Corkeron PJ, Nowacek DP, …, Kraus SD (2012) Evidence that ship noise increases stress in right whales. Proceedings of the Royal Society B: Biological Sciences, 279(1737), 2363–2368. https://doi.org/10.1098/rspb.2011.2429
Rosel PE, Tiedemann R, Walton M (1999) Genetic evidence for limited trans-atlantic movements of the harbor porpoise, Phocoena phocoena. Mar Biol (Berlin) 133(4):583–591
Rosel PE, France SC, Wang JY, Kocher TD (1999a) Genetic structure of harbour porpoise Phocoena phocoena populations in the northwest Atlantic based on mitochondrial and nuclear markers. Mol Ecol 8(s1):S41–S54. https://doi.org/10.1046/j.1365-294X.1999.00758.x
RStudio Team (2015) RStudio: Integrated Development for R. RStudio, Inc., Boston, MA. Retrieved from http://www.rstudio.com/
SAMBAH (2016) SAMBAH - Static Acoustic Monitoring of the Baltic Sea Harbour porpoise. LIFE Project Number LIFE 08 NAT/S/000261 European Commission, 77pp. available at http://www.sambah.org/SAMBAH-Final-Report-FINAL-for-website-April-2017.pdf. 1–77.
Santos M, Pierce GJ, Learmonth JA (2004) Variability in the diet of harbour porpoises (Phcoena phocoena) in Scottish waters 1992–2003. Mar Mamm Sci 20(January):1–27
Santure AW, Stapley J, Ball AD, Birkhead TR, Burke T, Slate J (2010) On the use of large marker panels to estimate inbreeding and relatedness: empirical and simulation studies of a pedigreed zebra finch population typed at 771 SNPs. Mol Ecol 19(7):1439–1451. https://doi.org/10.1111/j.1365-294X.2010.04554.x
Schaffeld T, Schnitzler JG, Ruser A, Woelfing B, Baltzer J, Siebert U (2020) Effects of multiple exposures to pile driving noise on harbor porpoise hearing during simulated flights—An evaluation tool. J Acoust Soc Am 147(2):685–697. https://doi.org/10.1121/10.0000595
Scheidat M, Gilles A, Kock KH, Siebert U (2008) Harbour porpoise Phocoena phocoena abundance in the southwestern Baltic Sea. Endanger Species Res 5(2–3):215–223. https://doi.org/10.3354/esr00161
Shafer ABA, Peart CR, Tusso S, Maayan I, Brelsford A, Wheat CW, Wolf JBW (2017) Bioinformatic processing of RAD-seq data dramatically impacts downstream population genetic inference. Methods Ecol Evol 8(8):907–917. https://doi.org/10.1111/2041-210X.12700
Siebert U, Wünschmann A, Weiss R, Frank H, Benke H, Frese K (2001) Post-mortem findings in harbour porpoises (phocoena phocoena) from the German North and Baltic seas. J Comp Pathol 124(2–3):102–114. https://doi.org/10.1053/jcpa.2000.0436
Siebert U, Tolley KA, Víkingsson GA, Ólafsdottir D, Lehnert K, Weiss R, Baumgärtner W (2006) Pathological findings in Harbour porpoises (Phocoena phocoena) from Norwegian and Icelandic Waters. J Comp Pathol 134(2–3):134–142. https://doi.org/10.1016/j.jcpa.2005.09.002
Siebert U, Blanchet MA, Teilmann J, Anderson Hansen K, Kristensen J, Bunskoek P, …, Desportes G (2020) Haematology and clinical blood chemistry in harbour porpoises (Phocoena phocoena) from the inner Danish waters. Environ Int 143(June):105937. https://doi.org/10.1016/j.envint.2020.105937
Sokal RR, Rohlf FJ (2012) Biometry: the principles and practice of statistics in biological research. (Ed.), W.H. Freeman and Company New York. FOURTH EDI). https://doi.org/10.2307/2412280Jerry Correa Marketing
Sommer RS, Pasold J, Schmölcke U (2008) Post-glacial immigration of the harbour porpoise (Phocoena phocoena) into the Baltic Sea. Boreas 37:458–464. https://doi.org/10.1111/j.1502-3885.2008.00034.x
Sonntag RP, Benke H, Hiby AR, Lick R, Adelung D (1999) Identification of the first harbour porpoise (Phocoena phocoena) calving ground in the North Sea. J Sea Res 41(3):225–232. https://doi.org/10.1016/S1385-1101(98)00050-1
Sveegaard S, Galatius A, Dietz R, Kyhn L, Koblitz JC, Amundin M, …, Teilmann J (2015) Defining management units for cetaceans by combining genetics, morphology, acoustics and satellite tracking. Global Ecol Conserv 3:839–850. https://doi.org/10.1016/j.gecco.2015.04.002
Tiedemann R, Harder J, Gmeiner C, Haase E (1996) Mitochondrial DNA sequence patterns of Harbour porpoises (Phocoena phocoena) from the North and the Baltic Sea. Z Für Säugetierkunde 61:104–111
Tiedemann R, Hardy O, Vekemans X, Milinkovitch MC (2000) Higher impact of female than male migration on population structure in large mammals. Mol Ecol 9(8):1159–1163. https://doi.org/10.1046/j.1365-294X.2000.00975.x
Tolley KA, Rosel PE (2006) Population structure and historical demography of eastern North Atlantic harbour porpoises inferred through mtDNA sequences. Marine Ecology Progress Series, 327(December 2013), 297–308. https://doi.org/10.3354/meps327297
Tolley KA, Víkingsson GA, Rosel PE (2001) Mitochondrial DNA sequence variation and phylogeographic patterns in harbour porpoises (Phocoena phocoena) from the North Atlantic. Conserv Genet 2(4):349–361. https://doi.org/10.1023/A:1012561811074
Unger B, Herr H, Benke H, Böhmert M, Burkhardt-Holm P, Dähne M, …, Siebert U (2017) Marine debris in harbour porpoises and seals from German waters. Mar Environ Res 130:77–84. https://doi.org/10.1016/j.marenvres.2017.07.009
Unger B, Baltzer J, Brackmann J, Brasseur S, Brügmann M, Diederichs B, …, van Neer A (2022) Marine mammals | Wadden Sea Quality Status Report. Wadden Sea Quality Status Report. Retrieved from https://qsr.waddensea-worldheritage.org/reports/marine-mammals
Van Cise AM, Baird RW, Baker CS, Cerchio S, Claridge D, Fielding R, …, Morin PA (2019) Oceanographic barriers, divergence, and admixture: Phylogeography and taxonomy of two putative subspecies of short-finned pilot whale. Mol Ecol 28(11):2886–2902. https://doi.org/10.1111/mec.15107
Varjopuro R, Andrulewicz E, Blenckner T, Dolch T, Heiskanen AS, Pihlajamäki M, …, Psuty I (2014) Coping with persistent environmental problems: systemic delays in reducing eutrophication of the Baltic Sea. Ecol Soc 19(4). https://doi.org/10.5751/ES-06938-190448
Viaud-Martínez KA, Vergara MM, Gol’din PE, Ridoux V, Öztürk AA, Öztürk B, …, Bohonak AJ (2007) Morphological and genetic differentiation of the Black Sea harbour porpoise Phocoena phocoena. Mar Ecol Prog Ser 338:281–294. https://doi.org/10.3354/meps338281
Víkingsson GA, Ólafsdóttir D, Sigurjónsson J (2003) Geographical, and seasonal variation in the diet of harbour porpoises (Phocoena phocoena) in Icelandic coastal waters. NAMMCO Sci Publications 5:243. https://doi.org/10.7557/3.2829
Viricel A, Pante E, Dabin W, Simon-bouhet B (2014) Applicability of RAD-tag genotyping for inter-familial comparisons: empirical data from two cetaceans. HAL Id: Hal-00908459. https://hal.archives-ouvertes.fr/hal-00908459
Walton MJ (1997) Population structure of harbour porpoises Phocoena phocoena in the seas around the UK and adjacent waters. Proceedings of the Royal Society B: Biological Sciences, 264(1378), 89–94. https://doi.org/10.1098/rspb.1997.0013
Waples RS, Hoelzel AR, Gaggiotti O, Tiedemann R, Palsbøll PJ, Cipriano F, …, Lang AR (2018) Guidelines for genetic data analysis. J Cetacean Res Manage 18:33–80
Wiemann A, Andersen LW, Berggren P, Siebert U, Benke H, Teilmann J, …, Tiedemann R (2010) Mitochondrial Control Region and microsatellite analyses on harbour porpoise (Phocoena phocoena) unravel population differentiation in the Baltic Sea and adjacent waters. Conserv Genet 11(1):195–211. https://doi.org/10.1007/s10592-009-0023-x
Wisniewska DM, Johnson M, Teilmann J, Rojano-Doñate L, Shearer J, Sveegaard S, …, Madsen PT (2016) Ultra-high foraging rates of Harbor Porpoises make them vulnerable to anthropogenic disturbance. Curr Biol 26(11):1441–1446. https://doi.org/10.1016/j.cub.2016.03.069
Wisniewska DM, Jonson M, Tielmann J, Siebert U, Galatius A, Dietz R, Madsen PT (2018) High rates of vessel noise exposure on wild harbour porpoises (Phocoena phocoena). Proceedings of the Royal Society B, 285, 20172314. https://doi.org/10.1098/rspb.2017.2314
Yu ZP, Liu X, Zhang B, Shan L, Seim I, Yang G, Xu SX (2022) High-quality chromosome-level genome assembly of the melon-headed whale (Peponocephala electra). Zoological Res 43(6):931–934. https://doi.org/10.24272/j.issn.2095-8137.2022.189
Yuan Y, Zhang P, Wang K, Liu M, Li J, Zheng J, …, Li S (2018) Genome sequence of the freshwater yangtze finless porpoise. Genes 9(4):1–9. https://doi.org/10.3390/genes9040213
Zhou X, Guang X, Sun D, Xu S, Li M, Seim I, …, Yang G (2018) Population genomics of finless porpoises reveal an incipient cetacean species adapted to freshwater. Nat Commun 9(1):1–8. https://doi.org/10.1038/s41467-018-03722-x
Acknowledgements
We thank Gísli Víkingsson and Þorvaldur Gunnlaugsson for organizing access to Icelandic porpoise samples. We thank Garry B. Stenson for providing access to Canadian porpoise samples. We thank Ljerka Lah for the support in ddRAD labwork. All samples were taken from carcasses, collected from local authorities holding permission granted by the respective officials. Large-scale computations were carried out on resources provided by Prof. Dr. Michael Lenhard at University of Potsdam and the High-Performance Computing Cluster Orson2, managed by ZIM (Zentrum für Informationstechnologie und Medienmanagement) at the University of Potsdam.
Funding
Open Access funding enabled and organized by Projekt DEAL. Financial support for part of the analyses came from the Bundesamt für Naturschutz (FKZ#3514824600). Further financial support came from the University of Potsdam.
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
R.T. designed and supervised the study; H.B., A.R., C.P., G.M.S., M.T.O, V.B., M.P.H.J., A.A.Ö., B.Ö., J.W.L. and U.S. provided samples and associated biological information. B.D.C. and K.H. performed molecular lab work. M.A. executed bioinformatic analyses and analysed the data, J.C. assisted with bioinformatics; M.A. and R.T. interpreted the results, M.A. wrote the draft manuscript with input from R.T. All authors edited and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
All work of Vincent Biard regarding this manuscript was performed at the University of Copenhagen and the Greenland Institute of Natural Resources.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Autenrieth, M., Havenstein, K., De Cahsan, B. et al. Genome-wide analysis of the harbour porpoise (Phocoena phocoena) indicates isolation-by-distance across the North Atlantic and potential local adaptation in adjacent waters. Conserv Genet 25, 563–584 (2024). https://doi.org/10.1007/s10592-023-01589-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-023-01589-0