Abstract
There has been a drastic decline in the number of eastern black rhinoceros (Diceros bicornis michaeli) across Africa, leaving individuals restricted to small, isolated populations that are vulnerable to extinction. Focusing on highly threatened populations in Tanzania, this study investigated the genetic impacts of past management interventions on mitochondrial control region diversity in extant subpopulations, assessed whether there has been evidence of dispersal of females between populations based on haplotype sharing, and related current haplotype diversity to historical patterns. Across extant subpopulations in Tanzania, six maternal haplotypes were identified, with an overall haplotype diversity of h = 0.72 but lower overall nucleotide diversity within populations (π = 0.017) compared to historical populations (π = 0.021). Translocated populations did not share haplotypes with native populations, even though all haplotypes from translocated individuals had been found among historic samples from Kenya. This suggests that translocations have been successful at restoring previous diversity to the region but that the current Intensive Protection Zone (IPZ) management practices have restricted the movement of females between subpopulations. Extant haplotypes were distributed among three East African haplogroups described in previous studies, suggesting that multiple lineages have been preserved despite the loss of historical haplotypes. Our recommendation is to enhance the utilisation of previous translocations by enabling the natural movements of individuals between subpopulations. Such a change in management strategy could be less costly both economically and in terms of animal welfare than the alternative of using genetic data to target specific animals for translocation in order to supplement diversity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding how animal populations vary within their environment is essential for developing effective conservation and managements plans; this becomes critical when dealing with endangered species. Incorporating genetic information into conservation management plans can help to reduce extinction risks by minimizing loss of genetic diversity through inbreeding, identifying populations of conservation concern, inferring population structure, resolving taxonomic uncertainties to define management units within species, detecting hybridization, defining sites for reintroductions, and choosing the best populations for reintroduction and forensics (Caughley 1994; Frankham 1995). There is, therefore, no doubt that the field of conservation genetics is key in efforts to attain sustainable biodiversity conservation.
The eastern black rhinoceros (Diceros bicornis michaeli; also known as the eastern hook-lipped rhinoceros) is a subspecies that was once widely distributed throughout South Sudan, Uganda, Ethiopia, Kenya and north-central Tanzania (Groves 1967; Hillman-Smith & Groves 1994). However, the population has declined by 90% in the last three generations, from an estimate of 70,000 individuals across Africa in the late 1960s to only 3800 in 1987, due to intensive poaching for their horns and habitat loss (Cumming et al. 1990). In Tanzania, the eastern black rhinoceros population had dropped from approximately 10,000 in the 1960s to only 46 by 1997 (Emslie & Brooks 1999). The few remaining individuals were restricted to a series of small and isolated populations (Makacha et al. 1982; Sinclair and Arcese 1995). By the 1990s in Tanzania, only three subpopulations remained: (1) three individuals in the Moru kopjes in the southern part of the Serengeti National Park; (2) 10 individuals in the Nyamalumbwa-Maasai Mara in the northern Serengeti-Mara ecosystem—a transboundary population between Kenya and Tanzania; and (3) 13 individuals in the Ngorongoro Crater. In 1997, eastern black rhinoceroses were first reintroduced to Tanzania. Two females were translocated from Addo Elephant National Park to the Ngorongoro Crater but their ancestors originally had been introduced to Addo Elephant National Park from Kenya in 1961 and 1962 (Hall-Martin 1984), so they were originally of east African origin. This was followed by further reintroductions to establish new populations in Mkomazi National Park and two additional populations in the Serengeti-Mara Ecosystem (Ndasiata and Ikorongo-Grumeti) from five captive populations, including: Port Lympne Wild Animal Park in the United Kingdom; Dvůr Králové Zoo in the Czech Republic; San Diego Zoo Safari Park in the United States of America; and Thaba Tholo private game ranch and Addo Elephant National Park in South Africa. Since then, both the native and reintroduced individuals have been managed in Intensive Protection Zones (IPZ) as separate subpopulations (Fyumagwa and Nyahongo 2010). IPZs are designed to provide an area of enhanced security by increasing protection and monitoring measures, such as specialized anti-poaching patrols, the use of advanced technology for identifying and tracking individual animals, and engaging local communities in conservation efforts. The only fences associated with IPZs are in Mkomazi and Grumeti; Mkomazi has a large, fenced enclosure to constrain eastern black rhinoceros to the IPZ, and Grumeti has a fence only on the western boundary of the protected area to minimise wildlife-livestock conflicts but is open on the eastern side that borders the Serengeti. The other four IPZs (Moru, Ngorongoro, Nyamalumbwa, and Maasai Mara) do not have physical barriers but animals are restricted within the subpopulations using GPS collars and geo-fencing technology (i.e. a virtual boundary). Rangers track movements of individuals within the IPZ and receive alerts when an animal moves out of their native zone. Eastern black rhinoceros outside the IPZ are pushed back into their designated area. However, this intensive protection strategy could come at a cost to natural dispersal.
Eastern black rhinoceros are solitary animals that establish and defend individual territories. While males are known for their territorial behavior and active defence of their range, females are more tolerant of each other's presence (Tatman et al. 2000). Female dispersal in this species is a behavioural pattern where individuals leave their birth area to establish their own territories and breeding opportunities (Reid et al. 2007). They may either stay near their natal range (philopatry) or move away to find vacant territories (dispersal). Dispersing females typically leave their natal area before reaching sexual maturity (around 3–6 years of age) and undertake movements covering significant distances, searching for unoccupied territories to establish their own home ranges (Hillman-Smith and Groves 1994). In the Serengeti, the average size of eastern black rhinoceros home ranges vary from approximately 40 to 133 square kilometres, whereas in the Ngorongoro area, it spans from 2.6 to 58.0 square kilometres (Frame 1980). Therefore, restricting dispersal could compromise genetic diversity, increase inbreeding, and reduce the spatial distribution of the eastern black rhinoceros populations in Tanzania.
As a result of past re-introductions, coupled with intensive protection and monitoring, the number of eastern black rhinoceroses in Tanzania has increased from 24 individuals in 1995 to 177 by the end of 2019 (TAWIRI 2019). Whilst this approach has yielded success in rehabilitating these closed subpopulations, the potential impacts of inbreeding depression are unknown because empirical genetic information was not considered in the selection of the founder individuals. The consequences of this demographic bottleneck on the genetic diversity for the small remote subpopulations could also result in additional impacts, including reduced viability of the population to evolve in response to extreme climates, parasitic burden or diseases epidemics (Gaines et al. 1997; Frankham et al. 2019).
Inbreeding can put populations at risk of extinction by increasing levels of homozygosity and exposing deleterious recessive alleles that could weaken reproductive fitness and ability to survive, resulting in inbreeding depression (Frankham 1995). Furthermore, by virtue of it being stronger than selection, genetic drift can cause unpredictable loss of adaptive alleles or retention of deleterious alleles (Hartl and Clark 1997). Small, isolated populations are often also characterised by restricted gene flow, as there is less chance of immigration and emigration (Frankham 1995). Apart from re-introductions from captive populations, translocation of wild individuals between different populations is another strategic management intervention (Fyumagwa and Nyahongo 2010). Such interventions are often used to balance the harmful effects of small population size and maintain natural evolutionary processes (Sinclair and Arcese 1995; Seddon et al. 2014). However, both reintroductions and translocations are only effective if the individuals being moved are sufficiently different from the host population to offset the effects of inbreeding (Jackson and Hobbs 2009). Therefore, genetic relatedness between the donor and the recipient populations is used as a key tool to inform suitability of different management interventions. It is for these reasons that establishing the current genetic health of the isolated sub-populations within Tanzania and that of the neighbouring cross-border population of Maasai Mara in Kenya becomes of paramount importance.
A pan-African assessment of the genetic status of black rhinoceros populations using microsatellite markers and mitochondrial DNA (mtDNA) sequencing revealed a 69% loss of mtDNA variation of the species (Moodley et al. 2017). Low genetic diversity and high inbreeding were also established in the Maasai Mara sub-population in Kenya compared to other larger subpopulations in a previous study (Muya et al. 2011). Across the entire species range, seven haplogroups have been identified, based on a combination of geographic distribution and phylogenetic clustering: WW, West Africa (west of the Shari-Logone River system); CV, Chari-Victoria (east of the Shari-Logone River to East Africa); NE, North-East Africa; EA, East Africa; CE, Central Africa (separated from EA by the Zambezi River); RU, Ruvuma region between Kilombero and Shire Rivers; and Southern Africa, which was subdivided further based on spatial distribution into Northern (SN), Eastern (SE) and Western (SW) lineages (Moodley et al. 2017). Most recently, de novo sequence analysis of genomes from all five extant and three extinct rhinoceros species has shown strong support of the geographical hypothesis of rhinoceros evolution and confirmed low genomic diversity in all extant rhinoceroses (Liu et al. 2021). However, none of these studies included representative samples from the current populations in Tanzania, so little is known about the genetic impacts of the severe population declines and subsequent management practices to increase numbers in this region. Thus, revealing the maternal diversity will help conservation efforts with regards to current management practices focused on translocations of individuals and to inform population viability assessments.
In order to compare current patterns of genetic variation in Tanzania with the widest geographic sampling from other studies, this study focused on the maternally inherited mtDNA control region. Our specific aims were to investigate: (1) the impacts of past translocations on diversity of maternal lineages in extant subpopulations; (2) whether there has been evidence of dispersal of females between populations based on haplotype sharing; and (3) how current haplotype diversity relates to historical patterns.
Materials and methods
Study area description
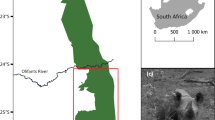
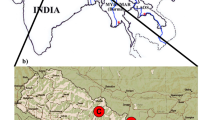
We sampled the East African subspecies of eastern black rhinoceros, D. b. michaeli, from the six extant protected subpopulations in Tanzania and one transboundary population in the Maasai Mara in Kenya (Fig. 1). Each subpopulation has had a different history of demographic changes and re-introduction strategies, as detailed below.
Maasai mara
The Maasai Mara Game Reserve in Kenya is located in the northern portion of the Serengeti-Mara ecosystem (Fig. 1). There were approximately 120 eastern black rhinoceros in 1971 but this number plummeted to 18 individuals by 1984 due to poaching (Moehlman et al. 1996). It is the only population in Kenya with free-ranging indigenous inhabitants unaffected by translocations (Muya et al. 2011). At present, there are 25 eastern black rhinoceros in this population (Table 1) that utilize areas across the border between Tanzania and Kenya. Although there are separate monitoring programmes on the two sides of the border, animals are known to move between the Maasai Mara IPZ and the Nyamalumbwa IPZ within the Serengeti National Park.
Nyamalumbwa
The Nyamalumbwa rhinoceros project works to conserve the eastern black rhinoceros inhabiting the cross-border area between northern Serengeti in Tanzania and the Maasai Mara National Reserve in Kenya (TAWIRI 2019). The project started in 1999, with only four pioneer native individuals; i.e. one male and three females (Fyumagwa and Nyahongo 2010). The Nyamalumbwa population moves freely across the international border and often interacts with the Maasai Mara population to the north and is adjacent to the re-introduced population at Ndasiata to the south. There are currently 20 individuals in the population (Table 1).
Ndasiata
The Ndasiata rhinoceros project (Serengeti Rhinoceros Repatriation Project) is situated in the north-eastern part of the Serengeti National Park (Fig. 1). The population was re-introduced in 2009, with the main objective to return indigenous animals to their native habitat. Five eastern black rhinoceros (two males and three females) were reintroduced from a captive population in Thaba Tholo, Thabazimbi, South Africa. The original animals in the Thaba Tholo captive population came from Tsavo National Park in Kenya and had been caught in 1961, during a period of high poaching (Hall-Martin 1984). The population has increased to 9 individuals (Table 1).
Moru kopjes
The Moru rhinoceros project strives to conserve eastern black rhinoceros inhabiting the southern part of the Serengeti National Park (Fig. 1). The population started with only one male and two females. While the two females were residents who survived poaching crises in the late 1980’s and early 1990’s, the male migrated from the Ngorongoro Crater in 1994 (Fyumagwa and Nyahongo 2010). The three founders successfully reproduced to generate 40 individuals in the current population (Table 1).
Ngorongoro crater
The Ngorongoro Conservation Area (NCA) occupies the southern side of the Serengeti-Mara ecosystem. Between 1964 and 1966, there were 108 eastern black rhinoceros in the NCA but, due to poaching in the 1990’s, only 10 individuals remained in the area. In 1997, two female eastern black rhinoceroses were introduced from Addo Elephant National Park in South Africa. The parental stock of these individuals were initially sourced from the Kibodo area in Kenya in 1961 and 1962 (Hall-Martin 1984). Currently, the NCA holds the largest population of free ranging eastern black rhinoceros in Tanzania (Table 1).
Grumeti
Grumeti-Ikorongo is a conservation area situated in the northern part of Tanzania, adjacent to the Serengeti National Park, and is part of the larger Serengeti-Mara ecosystem. The area includes the Grumeti Game Reserve and the Ikorongo Game Reserve. The Grumeti Rhino Reintroduction project initiative combined habitat restoration, anti-poaching measures, with the reintroduction of eastern black rhinoceros to the reserve from various sources. As of September 2021, the project had successfully reintroduced 18 individuals that had been maintained in captive conditions, from San Diego Zoo, Port Lympne Park in the UK and Thaba Tholo in South Africa.
Mkomazi
The Mkomazi Rhinoceros Sanctuary is in Mkomazi National Park. This is actually the southern extension of Kenya’s Tsavo West National Park ecosystem (Mbeyale and Songorwa 2008). Historically, eastern black rhinoceros would have moved between these two areas; however, fencing now restricts their movements (Homewood and Brockington 1999). The sanctuary was established in 1997 as a breeding ground for eastern black rhinoceros, with the aim of restoring a wild population. The starting population was composed of individuals from a collection of different zoos around the world: five from Addo Elephant National Park in South Africa; three from Dvur Kravole Zoo in the Czech Republic; and three from Port Lympne Wild animal Park, UK (Fyumagwa and Nyahongo 2010). The population currently has 30 individuals (Table 1).
Sample collection
For each subpopulation, samples were collected opportunistically during ear-notching operations designed to provide unique individual identification or during routine veterinary interventions. Samples include ear tissue, whole blood in EDTA and serum. In addition, biopsy darts were used to collect tissue samples from three young individuals from Mkomazi that had not yet been included in the ear notching campaigns (Table 1). Our strategy was to sample as many of the extant individuals as possible, rather than targeting particular individuals that may be related or occupy adjacent home ranges.
DNA extraction, mtDNA amplification and sequencing
Total genomic DNA from serum, EDTA blood or tissue samples was extracted using DNeasy® DNA kits following the manufacturer’s protocol (Qiagen Inc., Valencia, CA, USA, 2014). A 532 bp fragment of the mtDNA control region was amplified using the mt15996L (5′-TCCACCATCAGCACCCAAAGC-3′) and mt16502H (5′-TTTGATGGCCCTGAAGTAAGAACCA-3′) primers, as described by Brown and Houldon (2000). The primers target the D. b. michaeli mtDNA control region at positions 15,408 and 15,939 (Moodley et al. 2017).
Polymerase chain reactions were carried out in a 20 µl reaction containing 2 µl of DNA diluted to 1/100, 2 µl of 1 × PCR buffer, 1.2 µl of 50 mM MgCl2, 2 µl of 25 mM dNTP, 0.2 mg/µl purified BSA, 0.4 µl of each primer (10 µM), 0.2 µl of Taq polymerase (5 U/ µl) and 11.6 µl of purified water. Reactions were denatured at 95 °C for 5 min, followed by 45 cycles of 94 °C for 30 s, 60 °C for 1 min, 72 °C for 1 min and a final extension of 72 °C for 10 min. Amplified products were sent to the University of Dundee Sequencing Service for Sanger sequencing on an ABI 3730 automated sequencer; samples were sequenced in both directions using the PCR primers. The resultant sequences were manually cleaned and the contigs assembled using Sequencher version 4.5 (Gene Codes Inc; Ann Arbor, Michigan).
Impacts of past translocations on genetic variation
The sequences obtained from the samples collected from extant populations were aligned with one another and grouped into unique haplotypes using Sequencher 4.5 (Gene Codes Inc; Ann Arbor, Michigan). The identity of each unique haplotype was determined using a Basic Local Alignment Search Tool (BLAST) search against the National Centre for Biotechnology Information (NCBI) database. The sequences were aligned using Clustal Omega (Sievers et al. 2011) and manually optimised using Se-Al version 2.0 (Rambaut 2002, http://tree.bio.ed.ac.uk/software/seal/). Thereafter, the sequences were collapsed into unique haplotypes using DNAsp v6 (Rozas et al. 2017) and haplotype frequencies for each population were calculated (see Online Resource 1).
Relationships among the extant and historical haplotypes were visualized using a minimum spanning haplotype network generated with PopArt version 1.7 (Leigh and Bryant 2015). Branch lengths were scaled according to the number of mutations separating linked haplotypes in the network.
Genetic diversity of the mtDNA control region for the entire population, as well as for each subpopulation, was independently assessed by calculating haplotype diversity (h) and nucleotide diversity (π) in Arlequin version 3.5 (Excoffier and Lischer 2010). Haplotype diversity (h) is the probability that two randomly sampled haplotypes from a population will be different from one another (Nei 1987). Nucleotide diversity (π) is the average number of nucleotide differences per site between two DNA sequences across all possible pairs in the sample population (Nei 1987). To assess changes in diversity over time, we compared the values from the extant Tanzanian and Maasai Mara populations to the historical populations sampled by Moodley et al (2017), Thuo et al. (2019) and Muya et al (2011).
Differentiation between subpopulations in Tanzania
For the current Tanzanian mtDNA control region sequences, population structure was assessed using analysis of molecular variance (AMOVA) in Arlequin 3.5 (Excoffier and Lischer 2010). Population differentiation was further assessed using pairwise genetic distances between each population based on Fst.
Phylogenetic context of Tanzanian haplotypes
For comparative analysis, mtDNA D-Loop data from captive and wild black rhinoceros populations were obtained from GenBank. These sequences were deposited by Thuo et al. (2019), Moodley et al. (2017), Kotze et al. (2014), Muya et al (2011), Willerslev et al. (2009) and Brown and Houlden (2000) (see Online Resource 1). Only Moodley et al. (2017) included samples from Tanzania but two of the other studies were focused on samples from Kenya, allowing a broader context for the relative frequency of East African haplotypes; Thuo et al. (2019) provided 25 samples from Lake Nakuru National Park and Muya et al. (2011) included samples from 12 Kenyan subpopulations but only deposited unique haplotypes to Genbank. We used the ClustalW multiple alignment package in the BioEdit software version 7 (Hall, 2017, https://thalljiscience.github.io/page2.html) to align sequences obtained from the current study with a total of 444 other sequences retrieved from GenBank from these studies. The sequences were then collapsed into unique haplotypes and their frequencies recorded. The geographical region for each sample was identified (where that information was available; Online Resource 1) and each haplotype classified into one of the haplogroups identified by Moodley et al (2017). Where possible, haplotypes were further classified into either historical or modern groups (i.e., originating from museum archives, as opposed to being sampled from an extant population). No sampling dates were provided for the Muya et al. (2011) and Thuo et al. (2019) sequences but they were sampled from extant populations so they were considered as “modern”. Phylogenetic relationships among the haplotypes were analysed using black rhinoceros haplotypes recovered in our data set and from previous studies, with a white rhinoceros (Ceratotherium simum simum) sequence from GenBank as an outgroup (FJ004916.1; Online Resource 1). This analysis was done in BEAST v 2.5 (Bouckaert et al. 2019) under a Bayesian skyline model for lineage coalescence and the TN93 (Tamura and Nei 1993) nucleotide substitution model, as determined by model selection in the MEGA X software (Kumar et al. 2018). The analysis was run for 100 million MCMC steps, sampling the posterior distribution every 10,000 steps. The initial 10% of steps were discarded to ensure we sampled from the stationary part of the distribution. The final tree was visualised in Evolview software version 3 (Subramanian et al. 2019) and annotated using: the relative frequency of each haplotype, whether the haplotype was sampled from the extant (modern) or historical (museum samples) populations, the geographical regions of the haplotypes, and the haplogroups (WW, NE, CV, EA, CE, RU and South African) described by Moodley et al. (2017). For our samples and those lacking spatial data from other studies, we assigned haplogroups based on the positions in the phylogenetic tree (Online Resource 1). To further visualise relationships between the extant and historical haplotypes, we generated a minimum spanning haplotype network using PopArt version 1.7 (Leigh and Bryant 2015).
Results
mtDNA haplotype distribution
A total of 90 samples were sequenced successfully from Tanzania, with 12 more sequenced from the Maasai Mara in Kenya. The sequences included 25 polymorphic sites with no insertions or deletions and 438 monomorphic sites. Six haplotypes were found among the samples, which differed in frequency and distribution among the populations (Fig. 2). A comparison of the sampled mtDNA haplotypes with sequences in GenBank showed 100% similarity to published sequences for D. b. michaeli from Kenya for all except Haplotype 2, which most closely matched a sequence from Uganda that had been classified as D. b. ladoensis by the submitting authors (Table 2). Mkomazi had the highest number of haplotypes (four) while Maasai Mara had three and Ndasiata had a single haplotype. Haplotype 1 was found at the highest frequency and was shared among the five populations from Moru, Ngorongoro, Nyamalumbwa, Maasai Mara and Mkomazi. Haplotype 6 was shared among the three populations that were formed entirely from translocated individuals (Grumeti, Mkomazi and Ndasiata), but also a native population in Ngorongoro, which contains some translocated individuals. Haplotype 2 was found only in the native Moru and Maasai Mara populations. Haplotype 4 was restricted to Ngorongoro, Nyamalumbwa and Maasai Mara while haplotype 5 was found only in the Mkomazi and Grumeti populations (Fig. 2). The minimum spanning network showed that haplotype 2 had the highest number of mutations separating it from all others, whereas haplotypes 3 and 6 were separated by only one mutation. Haplotype 4 was separated from haplotypes 1 and 5 by the same number of mutations, forming a triangle (Fig. 3).
A minimum spanning network joining the six mtDNA control region haplotypes found in Tanzania. Circles represent haplotypes and the size is proportional to the haplotype frequency; ticks on branches show the number of mutations separating linked haplotypes; colours indicate the relative frequency of the haplotypes in each population
The overall mtDNA haplotype diversity across all extant eastern black rhinoceros sampled (n = 102) was 0.72, but the values varied considerably when each population was considered alone (Table 3). Mkomazi (n = 13) had the highest haplotype diversity (0.78), while Ndasiata had no haplotype diversity because it had only a single haplotype. Despite having only two haplotypes, Moru had the highest nucleotide diversity (π = 0.016), followed by Maasai Mara (π = 0.012); they shared the highly divergent haplotype 2 (Table 3).
The 19 haplotypes identified among 29 individuals sampled from historical populations in Tanzania by Moodley et al. (2017) included five of the haplotypes found in the current populations (Table 3). Haplotype 5 was not found among the historic samples from Tanzania but it had been identified among recent Kenyan samples (n = 4) and a Ugandan historic sample in Moodley et al. (2017). Of the three Tanzanian samples that Moodley et al. (2017) classified as “modern”, two had haplotype 4 and one had an additional haplotype not found in our extant samples (haplotype 44; Online Resource 1). Moodley et al. (2017) identified three haplotypes from eight “modern” individuals from the Maasai Mara (all collected in 1989); however, they did not find haplotype 2 in this population. Instead, they found an additional haplotype that was not found in the current samples analyzed by this study (haplotype 63; Online Resource 1). Haplotype diversity for Moodley’s historical samples from Tanzania was higher (h = 0.95) than for current populations in this study (h = 0.72; Table 3). The average nucleotide diversity across populations in the current study (π = 0.017) was less than the average of the previously described historical samples from Tanzania (π = 0.021).
Differentiation between subpopulations in Tanzania
For the comparison using AMOVA analyses, substantially more variation was explained within (60.2%) than among populations (39.8%), which may indicate lack of female migration between populations. However, comparison of pairwise Fst indicated substantial differentiation among individual subpopulations (Fig. 4), including between geographically proximate subpopulations such as Ndasiata and Nyamalumbwa.
Matrix of the pairwise Fst between subpopulations of eastern black rhinoceros in Tanzania. Fst values range from 0 for no differentiation to 1.0 for complete differentiation among subpopulations fixed for different alleles, with the intensity of colours in the heatmap showing higher differentiation. (Color figure online)
Phylogenetic context of Tanzanian haplotypes
Alignment to all available published sequences confirmed that all six of the Tanzanian haplotypes identified in this study had been found in other East African populations. Among the 146 sequences available from Kenya (including those newly sequenced here), 34 haplotypes were found; for samples classified by Moodley et al. (2017) nine haplotypes were identified only in historic samples (although they tended to be found in only a single individual), one (haplotype 42) only in samples classified as modern and nine were shared between time periods. The sequences from Muya et al. (2011) and Thuo et al. (2019) included 14 haplotypes that were not identical to any of those described in the Moodley et al. (2017) study. Haplotype 1 (haplotype 2 in Thuo et al. (2019)) was found at a substantially higher frequency in Kenya than all other haplotypes (n = 38; 26% of samples). The next most frequent were haplotype 33 (n = 15; not found in Tanzania) and haplotype 3 (n = 14; found only in the translocated populations Ndasiata and Mkomazi subpopulations in Tanzania). All three of these haplotypes were found in historic and recent samples from Kenya. As in our study, Moodley et al. (2017) found haplotypes 1, 2 and 4 in modern samples from the Maasai Mara population; Muya et al. (2011) also identified an additional 7 haplotypes (including our haplotypes 3 and 6). Haplotype 2 was also found in historical populations from Uganda. Haplotype 6, which was found in the populations that had been reintroduced (Ndasiata, Grumeti, Ngorongoro and Mkomazi) from South Africa and Europe, was also detected in Tanzanian historical populations and modern and historic Kenyan populations. Haplotype 5 was not detected in historical samples from Tanzania but was found in Kenyan modern populations and in a historic sample from Uganda that had been described as D. b. ladoensis.
Phylogenetic reconstruction of the mtDNA haplotypes using all published sequences (Fig. 5) showed three divergent lineages (using the classifications described in Moodley et al. (2017)), the most distinct of which (L1) comprised haplotypes sampled from West Africa (haplogroup WW from Nigeria and Cameroon). The second lineage (L2) was separable into two haplogroups: North-eastern (NE) and east of the Shari-Logone river system (Chari Victoria; CV). The last lineage (L3) is broadly distributed in eastern and southern sub-Saharan Africa and includes four haplogroups: EA, Eastern Africa; CE, Eastern Africa (Central); RU, Ruvuma (Eastern Africa South); and Southern Africa (including SN, SE and SW geographically defined lineages). The Tanzanian extant population haplotypes were mostly distributed into L3: haplotypes 1, 3, 5 and 6 in EA; and haplotype 4 in CE. However, the distinctive haplotype 2 was in the CV haplogroup from L2.
Bayesian phylogenetic tree of 79 mitochondrial DNA (mtDNA) control region haplotypes, obtained from a sample of 545 individual black rhinoceros sequences, with white rhinoceros (Ceratotherium simum simum) used as an outgroup. Branches with a posterior probability greater than 80% are indicated with a dot on the node. The relative frequency of each haplotype (proportional to the size of the circles) in various geographic regions (indicated with colours) is indicated to the right of the tree. Stars signify haplotypes from Tanzania, with the yellow circles indicating relative frequency in modern samples and peach circles historic. Relative frequency of haplotypes from other East African populations are indicated in various shades of green. Other East Africa includes Malawi, Ethiopia, Sudan, Eritrea and Somalia; Central Africa includes Cameroon, Chad, Nigeria and the Democratic Republic of Congo. Haplotypes from individuals sampled from Southern Africa are all indicated in red, combining lineages from: Southern Africa (Northern SN); Southern Africa (Eastern SE); and SW, Southern Africa (Western SW). Note that haplotypes that had been translocated to Tanzania from South African captive populations (haplotypes 3, 5 and 6) all were found among East African historic samples. Haplotypes for which their locations were not specified by the original authors are indicated by black dots (classified as unknown). (Color figure online)
Examining the relative frequency of extant East African haplotypes in a phylogenetic context (Fig. 5 and Fig. 6) clearly indicates a substantial loss of genetic diversity compared to historic samples, but the remaining haplotypes span multiple lineages within in the CE and EA haplogroups. The phylogenetic tree also confirms observational records that the animals that had been translocated from South Africa were originally of East African origin; however, the introduced haplotypes (3, 5, and 6) were all closely related and from a single EA cluster. None of the haplotypes at high frequency in the southern region of Africa (red circles, Fig. 5) were detected in the East Africa region. The tree and network also indicate that some of the diversity that has been lost in Tanzania has been found in modern samples from Kenya.
Minimum spanning network of 79 mitochondrial DNA (mtDNA) control region haplotypes, indicating the population groupings shown on Fig. 5. Each coloured circle denotes a haplotype, the size of each circle is proportional to the frequency at which that haplotype was observed in the data set, and the colours represent the country of origin of the samples. (Color figure online)
Discussion
This study presents the first assessment of diversity of mitochondria control region DNA of extant eastern black rhinoceros populations in Tanzania and a neighboring population in Maasai Mara, Kenya. The study adds to the assessment of the global distribution of mtDNA diversity described by Moodley et al. (2017) and provides critical information about maternal diversity that can inform conservation management of the current rhinoceros and other wildlife populations within the region. Although the comprehensive study by Moodley et al. (2017) had included three “recent” samples from Tanzania, only one was obtained from an animal which could still be alive. Therefore, our study fills a major gap in the knowledge about current maternal diversity of eastern black rhinoceros in two East African countries. As predicted from the drastic bottleneck that the extant populations experienced, with founding by only a few individuals, we found that current nucleotide diversity within the extant mtDNA control region was substantially lower than extant samples from Kenya and historical samples from Tanzania. This suggests that the current populations in Tanzania have lost genetic variation; therefore, more research utilising bioparental markers will be helpful to clearly assess the issue. In particular, the absence of haplotype sharing between the native and reintroduced populations may be influenced by the management strategies for Intensive Protection Zones, which limit the ability of rhinos to disperse naturally. This has likely impeded the potential beneficial effects of previous translocations. Although we found that recent translocations from captive populations have restored some of the former maternal lineages that were present historically, the IPZ strategy means that this only benefits the recipient population. Allowing animals to dispersal naturally across the greater Serengeti-Mara could spread the supplemented variation across populations at little cost, rather than relying only on more expensive and riskier managed translocations. In addition, some of the haplotypes that had been lost from Tanzania are still present in extant Kenyan samples; therefore, integrated cross-border management could provide a genetic “rescue” in both countries without introducing genetic variants from outside East Africa. We recognise that free movement could increase risks of poaching but animals already move outside of their IPZ regions and are forced to return; if dispersing animals were allowed to remain where they choose, this could allow better mixing without targeted interventions.
Current levels of genetic variation in Tanzania
Despite the recent decline in eastern black rhinoceros populations, moderate haplotype diversity (0.72) has been maintained, which is consistent with findings in other regional populations such as those in Zimbabwe (0.76) and Kenya (0.88) (Muya et al. 2011; Moodley et al. 2017). Haplotype diversity, nucleotide diversity and the number of haplotypes are all influenced by the proportion of the population sampled (Goodall-Copestake et al. 2012); in our study we sampled varying proportions of each subpopulation but we had the advantage of knowing how many maternal lineages were expected, due to detailed information on the founders. For example, we sampled all the individuals (40/40) of the current population from Moru but only two haplotypes (Haplotypes 1 and 2) were identified, which is consistent with founding from two females. Nevertheless, this population had the highest nucleotide diversity among the populations sampled (π = 0.017), which was comparable to pre-bottleneck historic patterns (π = 0.021), but was due to the large number of mutations separating haplotypes 1 and 2. In the Maasai Mara population, we found the same three haplotypes (haplotypes 1, 2 and 4; n = 12) as found by Moodley et al. (2017) with n = 15. However, the study conducted by Muya et al (2011; Fig. 2) found eight additional haplotypes in that population (including haplotypes 3 and 6); although they didn’t provide a detailed sample list, these appear to have been collected in the past decade, suggesting that diversity could be higher than our sampling suggested. We sampled all five individuals found in the other native subpopulation in this region (Nyamalumbwa) but identified only two haplotypes (haplotypes 1 and 4). Because haplotype 1 was the most frequent and is shared with several other native populations from Tanzania and Kenya (Moodley et al. 2017; Thuo et al. 2019), this suggests it could be an ancestral allele that reflects historical, rather than recent connectivity among these populations. Moodley et al. (2017) also sampled historic individuals from Zambia that had haplotype 1, despite being classified as a different subspecies (D. b. nyasae). Whether this reflects admixture between subspecies or misclassification would require further investigation.
The uneven distribution of haplotypes across our seven sampled populations means that allowing natural movements and dispersal between contiguous subpopulations (such as those in the greater Serengeti-Mara ecosystem) could enhance the genetic diversity. A similar approach has been suggested for bison herds in the USA and Canada in order to restore gene flow and enhance genetic diversity (Davies et al. 2022). An alternative approach could be to allow specific animals to move. For example, the relatively high presence of haplotype 1 could suggest over-representation of particular maternal lineages in the native populations (leading to increased inbreeding). Allowing animals that do not have haplotype 1 to move or to be translocated to populations where haplotype 1 is already present could be worthwhile and effective for restoring rarer haplotypes and lost genetic diversity. Despite retaining highly differentiated haplotypes, the concern for sustainability of the current populations is the low number of maternal lineages confirmed by the mtDNA variation. This means that allowing movements of native individuals might not be enough to maintain sufficient genetic diversity. For example, the Moru population was formed by three native eastern black rhinoceros, two females who survived the poaching catastrophe and one male which migrated from Ngorongoro. Moru has retained both maternal haplotypes from 40 sampled individuals, which is encouraging; however, it also illustrates the risk of inbreeding by maintaining isolated populations. The advantage of translocating animals is illustrated by Mkomazi, which had the highest number of haplotypes (four). Mkomazi had haplotypes that are shared with Grumeti, Ndasiata and Ngorongoro despite being isolated from the Serengeti-Mara ecosystem by a large geographical distance. Therefore, reintroducing animals from captive populations has clear advantages for enhancing genetic variation, and may be best utilized when animals from captive populations are reintroduced to populations that also include native individuals, such as Ngorongoro (Fyumagwa and Nyahongo 2010).
Differentiation between subpopulations in Tanzania
The AMOVA analysis revealed that a high percentage of variation exists among individuals within populations, but the overall differentiation was moderate, which could reflect historical sharing of alleles and movement between populations. In Kenya, Muya et al. (2011) found the highest Fst (0.729) between Chyulu and the Masai Mara, neither of which included introduced individuals. However, the Chyulu population had been bottlenecked to only two individuals, consistent with the presence of only two mtDNA haplotypes. Since the two populations are within the same ecosystem, this suggests recent restriction of movement, similar to our study. The lack of sharing of the introduced haplotypes in geographically close populations suggests that there is more restriction of maternal gene flow than home ranges would predict. Black rhinoceros are solitary and highly mobile; their estimated home ranges in the Serengeti are between 40 and 133 km2, regardless of sex (Frame 1980). However, the IPZ strategy, with no movement of individuals allowed between populations (Fyumagwa and Nyahongo 2010), means that these home range sizes are not realized. A vivid example is comparison between the Ndasiata and Nyamalumbwa subpopulations, which didn’t share any haplotypes and so had the highest Fst value (0.97), despite being located very close in the Serengeti-Mara ecosystem. This means that the advantages of the previous translocations to Ndasiata have not been extended to the native Nyamalumbwa population. Observational data suggests that the animals would move further if left more unconstrained. For example, on several occasions, individuals (especially bulls) from the Ngorongoro crater (NCA) have left their IPZ in search of new habitat or to escape from territorial fights; likewise, Moru individuals have escaped to Mwiba-Makoa areas. However, the management requires pushing them back into their respective IPZ. For example, in 2004 a young bull from NCA was sighted near lake Eyasi, 100 km away, but was immobilized and returned back (Fyumagwa and Nyahongo 2010). If individuals were allowed to naturally disperse, this could allow mixing of genetically distinct mating partners, without physical translocations.
Phylogenetic context of Tanzanian haplotypes
In Tanzania, translocation or assisted dispersal has been used as a tool for increasing the size of the eastern black rhinoceros population across the country. In previous years (1997–2018), a total of 23 individuals were translocated to Tanzania from areas outside East Africa but this was done without consideration of genetic variation (Fyumagwa & Nyahongo 2010). Only four haplotypes (haplotypes 1, 3, 5 and 6) were found from reintroduced eastern black rhinoceros sampled in this study. The parental stock of these individuals were captured from the Kibodo area, Tsavo National Park, Isiolo and Tana River in Kenya between 1960 and 1980 and taken outside East Africa to highly protected areas such as zoos and closed sanctuaries as a measure to rescue them from poaching in the wild (Hall-Martin 1984). This was confirmed by phylogenetic reconstruction of the mtDNA, which demonstrated that the maternal lineages introduced to Ndasiata, Mkomazi and Ngorongoro were of East African origin, despite individuals being translocated from European zoos or captive population in South Africa. However, three of the introduced haplotypes were closely related and clustered together on the tree, suggesting that the previous translocations achieved limited augmentation of genetic diversity in the extant populations. Furthermore, the presence of a wide range of the “lost” Tanzanian haplotypes in modern Kenyan samples (Muya et al. 2011; Moodley et al. 2017; Thuo et al. 2019) suggests that translocations within East Africa could be more beneficial, less costly, and less risky than long-distance translocations from Europe and South Africa.
The phylogenetic tree illustrates that mitochondrial control region sequence variation is highly structured, suggesting that careful consideration of which lineages to reintroduce could be beneficial (Fig. 5). Tanzanian extant populations haplotypes were distributed into all three haplogroups (CV, CE and EA) found in East Africa but there were notable gaps in the presence of particular clades that were present historically. The next step for specifically identifying individuals for translocation will be to assess the nuclear genome, not only to confirm the status of individuals with rare maternal haplotypes, but also to identify paternal contributions to the genetic diversity. We are currently taking a whole genome sequencing approach to address this question, based on a subset of the individuals used in this study.
Conservation implications
Our study has shown that the Tanzanian eastern black rhinoceros populations have lost substantial variation in the mtDNA from the recent population decline but still maintain moderate genetic diversity across the subpopulations. Recently translocated populations (such as Mkomazi and Ndasiata) have restored some of haplotypes that were previously present; however, the genetic benefit of translocation has been under-realized because animals are not permitted to move between Intensive Protection Zones (IPZ). Based on our results we recommend a combined management approach: (i) subpopulations that occur in the same ecosystem (such as the greater Serengeti-Mara ecosystem) should be managed as a single metapopulation rather than isolated IPZs. This would allow movement of individuals between regions (such as Ngorongoro to Maasai Mara) and could enhance supplementation of the native populations with additional genetic variation from past translocations; (ii) for populations that do not occur in the same ecosystem, such as Mkomazi, targeted reintroductions based on genetic variation may be the most effective way to reduce the effects of inbreeding and maintain genetic diversity; (iii) we recommend whole genome sequencing of nuclear DNA to further inform which individuals to translocate because this would provide evidence of both maternal and paternal contributions to genetic diversity; and finally (iv) we recommend that translocated animals be selected from extant populations within East Africa because there appears to be plenty of genetic variation in the mitochondrial DNA in these populations that were once historically connected. This also could be important to avoid translocation catastrophes, which can occur when animals that have been kept under benign captive conditions are released to wild environments (Chipman et al. 2008).
Data availability
The six unique haplotypes generated during the current study are available in the GenBank repository or on reasonable request from the corresponding author, accession numbers: OQ095383-OQ095388.
References
Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N (2019) BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS computational biology 15:e1006650
Brown SM, Houlden BA (2000) Conservation genetics of the black rhinoceros (Diceros bicornis). Conserv Genet 1:365–370. https://doi.org/10.1023/a:1011579807460
Caughley G (1994) Directions in conservation biology. J Anim Ecol 63:215–244. https://doi.org/10.2307/5542
Chipman R, Slate D, Rupprecht C, Mendoza M (2008) Downside risk of wildlife translocation. Dev Biol (basel) 131:223–232
Cumming, DHM, du Toit, RF, Stuart, SN, Cumming, DHM, du Toit, RF, Stuart, SN (1990) African elephants and rhinos. Status survey and conservation action plan. In: African elephants and rhinos Status survey and conservation action plan, pp. i-iv, 1–72. IUCN, Gland,
Davies G, McCann B, Jones L, Liccioli S, Penedo MC, Ovchinnikov IV (2022) Genetic variation of the mitochondrial DNA control region across plains bison herds in USA and Canada. PLoS One 17:e0264823. https://doi.org/10.1371/journal.pone.0264823
Emslie, R, Brooks, M (1999) African rhino: status survey and conservation action plan. Gland and Cambridge, UK IUCN/ SSC African Rhino Specialist Group,
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Frame GW (1980) Black rhinoceros (Diceros bicornis L.) sub-population on the Serengeti plains Tanzania. African Journal of Ecology 18:155–166. https://doi.org/10.1111/j.1365-2028.1980.tb00639.x
Frankham R (1995) Conservation genetics. Annu Rev Genet 29:305–327. https://doi.org/10.1146/annurev.ge.29.120195.001513
Frankham R, Ballou JD, Ralls K, Eldridge MDB, Dudash MR, Fenster CB, Lacy RC, Sunnucks P, Frankham R, Ballou JD, Ralls K, Eldridge MDB, Dudash MR, Fenster CB, Lacy RC, Sunnucks P (2019) Genetic problems in small isolated-populations and their remedies. Oxford Univ Press, New York
Fyumagwa RD, Nyahongo JW (2010) Black rhino conservation in Tanzania: translocation efforts and further challenges. Pachyderm 47:59–65
Gaines MS, Diffendorfer JE, Tamarin RH, Whittam TS (1997) The effects of habitat fragmentation on the genetic structure of small mammal populations. J Hered 88:294–304. https://doi.org/10.1093/oxfordjournals.jhered.a023107
Goodall-Copestake WP, Tarling GA, Murphy EJ (2012) On the comparison of population-level estimates of haplotype and nucleotide diversity: a case study using the gene cox1 in animals. Heredity 109:50–56. https://doi.org/10.1038/hdy.2012.12
Groves CP (1967) Geographic variation in the black rhinoceros, Diceros bicornis (L, 1758). Zeitschrift Fur Saugetierkunde 32:267–276
Hall-Martin A (1984) Kenya’s black rhinos in Addo, S. Africa. Newsletter of the African Elephant and Rhino Group 3:1–11. https://doi.org/10.2307/3504292
Hartl DL, Clark AG (1997) Principles of population genetics. Sinauer Associates, Sunderland, Massachusetts
Hillman-Smith AK, Groves CP (1994) Diceros Bicornis. Mammalian Species 455:1–8
Homewood K, Brockington D (1999) Biodiversity, conservation and development in Mkomazi Game Reserve, Tanzania. Glob Ecol Biogeogr 8:301–313. https://doi.org/10.1046/j.1365-2699.1999.00144.x
Jackson ST, Hobbs RJ (2009) Ecological restoration in the light of ecological history. Science 325:567–569. https://doi.org/10.1126/science.1172977
Kotze A, Dalton DL, du Toit R, Anderson N, Moodley Y (2014) Genetic structure of the black rhinoceros D( iceros bicornis) in south-eastern Africa. Conserv Genet 15:1479–1489. https://doi.org/10.1007/s10592-014-0632-x
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Leigh JW, Bryant D (2015) POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116. https://doi.org/10.1111/2041-210x.12410
Liu SL, Westbury MV, Dussex N, Mitchell KJ, Sinding MHS, Heintzman PD, Duchene DA, Kapp JD, von Seth J, Heiniger H, Sanchez-Barreiro F, Margaryan A, Andre-Olsen R, De Cahsan B, Meng GL, Yang CT, Chen L, van der Valk T, Moodley Y, Rookmaaker K, Bruford MW, Ryder O, Steiner C, Bruins-van Sonsbeek LGR, Vartanyan S, Guo CX, Cooper A, Kosintsev P, Kirillova I, Lister AM, Marques-Bonet T, Gopalakrishnan S, Dunn RR, Lorenzen ED, Shapiro B, Zhang GJ, Antoine PO, Dalen L, Gilbert MTP (2021) Ancient and modem genomes unravel the evolutionary history of the rhinoceros family. Cell 184:4874–4885. https://doi.org/10.1016/j.cell.2021.07.032
Makacha S, Msingwa MJ, Frame GW (1982) Threats to the Serengeti herds. Oryx 16:437–444. https://doi.org/10.1017/s0030605300018111
Mbeyale GE, Songorwa AN (2008) Conservation for whose benefit ? Challenges and opportunities for management of Mkomazi Game Reserve, Tanzania. People, protected areas and global change: participatory conservation in Latin America, Africa, Asia and Europe Perspectives of the Swiss National Centre of Competence in Research (NCCR) North-South. University of Bern, Geographica Bernensia 3:221–251
Moehlman PD, Amato G, Runyoro V (1996) Genetic and demographic threats to the black rhinoceros population in the Ngorongoro Crater. Conserv Biol 10:1107–1114. https://doi.org/10.1046/j.1523-1739.1996.10041107.x
Moodley Y, Russo IM, Dalton DL, Kotze A, Muya S, Haubensak P, Balint B, Munimanda GK, Deimel C, Setzer A, Dicks K, Herzig-Straschil B, Kalthoff DC, Siegismund HR, Robovsky J, O’Donoghue P, Bruford MW (2017) Extinctions, genetic erosion and conservation options for the black rhinoceros (Diceros bicornis). Sci Rep 7:41417. https://doi.org/10.1038/srep41417
Muya SM, Bruford MW, Muigai AWT, Osiemo ZB, Mwachiro E, Okita-Ouma B, Goossens B (2011) Substantial molecular variation and low genetic structure in Kenya’s black rhinoceros: implications for conservation. Conserv Genet 12:1575–1588. https://doi.org/10.1007/s10592-011-0256-3
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York. https://doi.org/10.7312/nei-92038
Reid C, Slotow R, Howison O, Balfour D (2007) Habitat changes reduce the carrying capacity of Hluhluwe-Umfolozi Park, South Africa, for critically endangered black rhinoceros Diceros bicornis. Oryx 41:247–254. https://doi.org/10.1017/s0030605307001780
Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sanchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol 34:3299–3302. https://doi.org/10.1093/molbev/msx248
Seddon PJ, Griffiths CJ, Soorae PS, Armstrong DP (2014) Reversing defaunation: restoring species in a changing world. Science 345:406–412. https://doi.org/10.1126/science.1251818
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Sinclair ARE, Arcese P (1995) Serengeti II: dynamics, management, and conservation of an ecosystem. University of Chicago Press, Chicago. https://doi.org/10.2307/5794
Subramanian B, Gao SH, Lercher MJ, Hu SN, Chen WH (2019) Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res 47:W270–W275. https://doi.org/10.1093/nar/gkz357
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial-DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. https://doi.org/10.1093/oxfordjournals.molbev.a040023
Tatman SC, Stevens-Wood B, Smith VBT (2000) Ranging behaviour and habitat usage in black rhinoceros, Diceros bicornis, in a Kenyan sanctuary. Afr J Ecol 38:163–172. https://doi.org/10.1046/j.1365-2028.2000.00235.x
TAWIRI (2019) Conservation and management plan for black rhino in Tanzania (2019 - 2023), 4th Edition. TAWIRI, Arusha, Tanzania,
Thuo DN, Kamau JM, Kariuki MW, Kibegwa FM, Kimiti K, Amimo JO, Githui EK (2019) Genetic assessment of a breeding population of black rhinoceros in Kenya using mitochondrial DNA D-loop sequencing. Genet Mol Res 18:1–11. https://doi.org/10.4238/gmr18203
Willerslev E, Gilbert MTP, Binladen J, Ho SYW, Campos PF, Ratan A, Tomsho LP, da Fonseca RR, Sher A, Kuznetsova TV, Nowak-Kemp M, Roth TL, Miller W, Schuster SC (2009) Analysis of complete mitochondrial genomes from extinct and extant rhinoceroses reveals lack of phylogenetic resolution. BMC Evol Biol 9:95. https://doi.org/10.1186/1471-2148-9-95
Acknowledgements
We are grateful to the following institutions for making their collections available to this study: Tanzania National Parks (TANAPA), Tanzania Wildlife Authority (TAWA), Ngorongoro Conservation Authority (NCA) and the Kenyan Wildlife Service (KWS). We thank the Nelson Mandela African Institute of Science and Technology, Arusha, Tanzania and the Kenyan Wildlife Services for providing us a platform in their laboratories to perform DNA extraction of the samples. A special thank you to Mike Bruford, who provided support and advice during the planning phases of the project; his untimely passing is an enormous loss to the conservation genetics community. Also, we thank the Tanzania Wildlife Research Institute (TAWIRI) and Tanzania Commission for Science and Technology (COSTECH) for granting us permits to conduct the study (Permit number 2021-449-NA-2021-150).
Funding
This work was funded by Zurich Zoo, the Paul Tudor Jones Family Trust. Ronald V. K. Mellya also received research support from a National Geographic early career grant [Grant number EC-51221R-18].
Author information
Authors and Affiliations
Contributions
The study conception and design were done by RVKM. Material preparation was performed by JGCH, BKM and RVKM. Sample collection was carried out by RVKM, DW, ESM, LK, MO, ISC and EME. Molecular analyses were performed by RVKM, EK and BKM. The first draft of the manuscript was written by RVKM and all authors commented on previous versions of the manuscript. All authors read and accepted the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mellya, R.V.K., Hopcraft, J.G.C., Eblate, E.M. et al. Mitochondrial DNA diversity of the eastern black rhinoceros (Diceros bicornis michaeli) in Tanzania: implications for future conservation. Conserv Genet 24, 905–919 (2023). https://doi.org/10.1007/s10592-023-01545-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-023-01545-y