Abstract
The status of many amphibian populations remains unclear due to undetected declines driven by disease and difficulties in obtaining accurate population estimates. Here, we used genome complexity reduction-based sequencing technology to study the poorly understood Littlejohn’s treefrog, Litoria littlejohni across its fragmented distribution in eastern Australia. We detected five identifiable genetic clusters, with moderate to strong genetic isolation. At a regional scale, population isolation was likely driven by population crashes, resulting in small populations impacted by founder effects. Moderate genetic isolation was detected among populations on the Woronora Plateau despite short distances between population clusters. Evidence of recent declines was apparent in three populations that had very small effective population size, reduced genetic diversity and high inbreeding values. The rates of inbreeding detected in these populations combined with their small size leave these populations at elevated risk of extinction. The Cordeaux Cluster was identified as the most robust population as it was the largest and most genetically diverse. This study exemplifies the value of employing genetic methods to study rare, cryptic species. Despite low recapture rates using traditional capture-recapture demographic methods, we were able to derive population estimates, describe patterns of gene flow, and demonstrate the need for urgent conservation management.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Amphibians are the most threatened vertebrate group in the world, with many species declining or facing extinction (Stuart et al. 2004; Mendelson et al. 2006). Frogs are threatened by a wide range of processes including climate change, pollution, habitat alteration and introduced species (Laurance 2008; Alton & Franklin 2017; Hayes et al. 2010; Cushman 2006; Kats & Ferrer 2003). Infectious disease is the greatest threat to amphibians since chytridiomycosis has caused the decline of over 500 species globally (Bower et al. 2017; Scheele et al. 2019). This disease is caused by the chytrid fungus, Batrachochytrium dendrobatitidis, and at least 43 frog species in Australia have declined in population size due to this pathogen (Berger et al. 1998; Skerratt et al. 2007; Scheele et al. 2017; Scheele et al. 2019). Long term population trajectories are highly variable for Australian frogs threatened by chytrid. Some species continue to decline while others appear to be stabilizing or recovering. For other Australian species, the severity of initial declines and changes to distribution in temperate Australia remain unclear due to a lack of base-line information (Scheel et al. 2017). It is also not clear to what extent poor monitoring of species has caused some declining populations to appear stable (Scheele et al. 2017). With so much uncertainty, there is a pressing need to obtain accurate estimates of frog population sizes and estimate gene flow for poorly monitored threatened species.
Within conservation research, DNA sequencing technologies are becoming increasingly popular as these technologies enhance and complement traditional population monitoring techniques (Hohenlohe et al. 2021). Genetic methods typically require lower sampling effort and can provide insight into behaviours, such as dispersal and breeding, making them highly suitable for species that are difficult to observe (Amato et al. 2009; Wang et al. 2009; McCartney-Melstad & Shaffer. 2015). In recent years, nuclear genome wide sequencing to identify single nucleotide polymorphisms (SNPs) have been used to greatly expand our knowledge of population connectivity, dispersal patterns, effective population sizes, mating systems and genetic diversity for several rare and cryptic species (Allendorf et al. 2010; Bradford et al. 2018; Harrison et al. 2019; Miller et al. 2018). By employing these methods alongside traditional fieldwork, researchers can develop more specific and informed conservation actions for species facing extinction (Frankham et al. 2019).
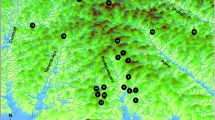
Littlejohn’s treefrog (Litoria littlejohni) is one Australian amphibian of conservation concern for which population dynamics are uncertain. This moderate sized (adult body length 42 mm) brown tree frog occurs in the temperate climatic region of south-east Australia. Recently, L. littlejohni was redefined and taxonomically split into two species, and subsequently its conservation status was assessed as Endangered (EN) (International Union for the Conservation of Nature Red List criteria, IUCN 2012) by Mahony et al. (2020). What is now considered L. littlejohni is found in three isolated regions in the state of New South Wales (NSW) (Mahony et al. 2020). The geographic range is restricted to the Sydney Basin bioregion (Thackway & Cresswell 1995) with apparently isolated populations on the Woronora Plateau in the southeast, to the Blue Mountains in the west, and to the Central Coast Range (Watagan Mountains) in the northeast (Fig. 1). It is unknown whether the populations found in the Blue Mountains, Central Coast Range and Woronora Plateau are connected or isolated from one another. Furthermore, the degree of gene flow between occupied sites within these populations is unknown.
a GPS points for all L. littlejohni and L. watsoni samples included in population genetic analysis represented by circles and triangles, respectively. b Zoomed in map of Woronora plateau showing river catchments in black lines, creeks in grey and sampled L. littlejohni individuals as triangles c Map of Woronora Plateau showing major roads in dark grey and black, sampled L. littlejohni individuals in triangles. CCR- Central Coast Range (maroon), BM—Blue Mountains (green), OH—O’Hare’s Catchment (dark purple), CAT—Cataract Catchment (orange), AVON—Avon Catchment (dark blue), CORD—Cordeaux Catchment (blue), NEP—Nepean Catchment (light purple), LW—Litoria watsoni (pink)
For species, such as L. littlejohni, that have a fragmented distribution across their range, it is important to consider patterns of gene flow and genetic processes such as Isolation by Distance and Isolation by Environment. Isolation by Distance refers to the process in which genetic differences increase with geographical distance as dispersal tends to decrease with distance (Wright 1943). Populations can also become more genetically distinct due to increases in environmental differences independent of geographical distance, which is termed Isolation by Environment (Wang & Summers 2010; Bradburd et al. 2013; Sexton et al. 2014; Wang & Bradburd 2014). When gene flow is restricted between populations, several genetic changes can occur in the resulting isolated populations, including reductions in effective population size, loss of genetic variation and increased local inbreeding within the isolated population (Frankham 2010). Geographically isolated populations are more likely to be vulnerable to stochastic environmental events including drought and fire, and stochastic demographic events including non-random mating, thus experiencing higher extinction risk (Frankham 2010).
The rarity of L. littlejohni has presented a scientific quandary. Determining the cause of rarity requires an understanding of population sizes and dynamics, which is difficult when little is understood about the species. Typically, L. littlejohni is found in dry sclerophyll or heath forests that are widespread community types across the east coast of NSW and the precise vegetation requirements remain unknown (White et al. 1994; Lemckert 2010; Klop-Toker et al. 2021). Additionally, exact breeding preferences remain poorly understood (Klop-Toker et al. 2021). It has only recently been confirmed that breeding peaks in late winter to early spring (daily mean minimum 6 °C and maximum 18 °C on Woronora plateau) (Gill et al. 2021). This poor understanding of habitat preferences and calling behaviours is thought to have led to mis-matched survey efforts and false absences in historical surveys conducted across the range (Gill et al 2021). Populations of L. littlejohni also do not appear to be large and densities at breeding ponds are relatively low compared to other Australian tree frogs, resulting in low capture rates at occupied sites (Mahony et al. 2020; Klop-Toker et al. 2021; Gillespie et al. 2016). Whether this low density is due to inherent low population density or due to declines is unknown (Lemckert 2004; Mahony et al. 2020). Litoria littlejohni is susceptible to chytrid with prevalence levels approximately 30% across all L. littlejohni populations (Klop-Toker et al. 2021), but there is no pre-chytrid population data for this species. There is evidence of gradual declines, in individual populations, particularly in the Blue Mountains where only three breeding locations on the King’s Tablelands have been confirmed in the past decade despite intensive survey efforts (Mahony et al. 2020). While there is evidence of L. littlejohni declines, there has been no formal estimate of population size for any population.
In this study, we aimed to use genetic data (SNPs) to 1) elucidate patterns of gene flow across fragmented populations, 2) determine the size of L. littlejohni populations 3) estimate genetic diversity and inbreeding within populations, and 4) outline priority conservation and research actions to promote long-term persistence of L. littlejohni.
Material and methods
Sample Collection and Selection
We conducted surveys for L. littlejohni between May 2018 and June 2021 in three regions of NSW that span the known distribution of the species—the Blue Mountains, Central Coast Range, and Woronora Plateau (Fig. 1). Within the Woronora Plateau, 18 × 100 m transects were distributed across four water catchments—O’Hare’s Creek, Cordeaux River, Avon River, and the Cataract River (Fig. 1). Within the Blue Mountains and Central Coast Range, we established two and nine sites respectively at fire dams or large ponds, rather than 100 m transects. Ponds at these two locations were selected instead of stream transects because L. littlejohni have been detected more commonly at dams and ponds in these two regions and were therefore considered more reliable sites for catching adult frogs or tadpoles.
Surveys were conducted using standardized methods across all seasons with the total number at a site ranging from one to 13 occasions. Sites with low survey occasions were typically established late into the project upon discovery of new occupied sites. Surveys for adults were conducted at night and involved a timed active search of stream/pond bank habitat aided by spotlights. This initial search was followed by a repeated active search in conjunction with call-playback to help locate male frogs. All adults caught were microchipped to reduce the likelihood of double sampling in future surveys and to allow the collection of recapture data. A hand-held GPS was used to collect the precise location (latitude and longitude) of frog collection to the nearest 5 m or the latitude and longitude of the site. We collected genetic samples by taking a 4 mm biopsy from the rear foot webbing of frogs with a snout to vent length (SVL) over 40 mm and stored samples in 70% ethanol.
We also collected genetic samples and GPS locations from L. littlejohni found opportunistically outside of surveyed transects or ponds to boost our sample size and geographical reach. During the day we visually inspected ponds for tadpoles at each site. When tadpoles were detected, we collected a subset of tadpoles by dip-net and tail tips were collected for genetic analysis. Tail tips were only collected from tadpoles longer than 18 mm in body length. Tadpole samples were included in genetic analysis if they were from sites where the adult sample size was low (< 6). We selected tadpole samples from different ponds along 100 m transects to reduce the likelihood of sequencing related individuals. Additionally, we included 26 samples that were collected and sequenced by Mahony et al. (2020) to extend the geographic reach and increase sample size. The main difference in methodology between the sample collection of Mahony et al. (2020) and that of the present study was the addition of microchipping adults in the present study (Table S2). While most of the samples from Mahony et al. (2020) came from eight additional sites, we included samples from three sites also surveyed during the present study to boost sample size at the locations. Overall, samples were collected from 37 locations—15 in Cordeaux River, 9 in the Central Coast range, 6 in the O’Hare’s Creek, 3 in the Blue Mountains, 2 site in Avon River, 1 in the Nepean River and 1 in the Cataract River (Fig. 1). Additionally, we included two double blinds to help detect whether we double sampled individuals due to microchip loss.
Litoria watsoni is the most closely related species to L. littlejohni (Mahony et al. 2020) and we used 12 Litoria watsoni samples from the NSW range as an outgroup. These samples were collected across 5 national parks between 2018 and 2020 using the same methods as described for L. littlejohni or were collected as per Mahony et al. 2020 (Fig. 1; Table S1). All genetic samples were stored in 70% ethanol.
DNA sequencing and initial filtering
Samples were sent to Diversity Arrays Technology Pty Ltd (Canberra, Australia) for DNA extraction following methods outlined in Kilian et al. (2012). SNP genotyping and discovery was carried out by Diversity Arrays Technology using the DArTseq protocol (Georges et al. 2018). Some of the major advantages of the DartSeq method is that it can handle lower DNA input and DNA quality than other sequencing methods. This sequencing method does not require a reference genome allowing for poorly understood species (non-model animal and plant species), such as our target L. littlejohni, to be studied and is therefore also cost effective. Briefly, this method employs PstL and Sphl restriction enzymes, custom proprietary bar-coded adapters, PCR amplification, and sequencing using Illumina HiSeq2500 following the methods detailed in Georges et al. (2018). Sequences generated were processed using a proprietary DArT analytical pipeline described by Georges et al. (2018). Samples from this study were co-analysed with Mahony et al. (2020) to ensure that the datasets could be merged.
The DArTSeq sequencing, and filtering pipelines provided a total of 62,368 SNPs. SNPs were further filtered using the DartR V2.3 package in Rstudio V2022.7.0.548 to obtain the highest quality data (Mijangos et al. 2022; RStudio Team 2022). We used gl.filter.locmectric to filter the read depth for both the reference and alternative allele to > 7. We then filtered SNPs to fit the following criteria: reproducibility of > = 99%, no monomorphic loci, SNP call rate > 97% (SNPs not found in 3% of the individuals), and no secondary SNPs. Secondary SNPs are those found within the same sequencing fragment and are likely to be linked (Gruber et al. 2018). Secondaries were removed by filtering out all but the first SNP with the same CloneID. We also excluded individuals with an individual call rate of < 95%, (i.e., individuals with > 5% of SNPs missing). We filtered for minor a frequency using a threshold of 0.00169 to remove overall rare alleles from the dataset.
After initial filtering for SNP quality, we assessed whether there were closely related individuals and double sampled individuals (due to microchip loss) that needed to be removed from the dataset. Related individuals and double sampling of individuals can heavily bias population structure as models typically assume individuals to be unrelated (Patterson et al. 2006; Anderson and Dunham 2008; O’Connell et al. 2019). To conduct this analysis, we created 5 small datasets based on sample region to help easily identify related individuals (Blue Mountains, Central Coast Range, Cordeaux/Avon, O’Hare’s/Cataract and L. watsoni). For each of these small datasets, we ran the initial filtering steps and then employed the bitwise.dist function from the poppr V2.9.3 R package suing default settings. This function calculates pairwise genetic distances between samples (Kamvar et al. 2014, 2015). We used the double-blind samples to calculate a threshold value for genetic distances. For pairs that differed by less than 0.01%, we removed one of the individuals from our overall data set. Once double sampled individuals were removed, we refiltered the data and we used a genetic relatedness network to assess relationships. We employed the gl.grm.network function from DartR, which uses a similar approach to that used by Goudet et al. (2018). This method is suited to populations for which little is known as it uses the kinship of all pair uses the average inbreeding coefficient as a reference value and is then subtracted from the inbreeding coefficient of each pair of distinct individuals (Goudet et al. 2018). We removed individuals so that no pair of individuals had a relatedness factor ≥ 0.25. This threshold was selected as sibling or parent–offspring relationships have a kinship value around 0.25 (Speed & Balding 2015). When all regions had been assessed, we made two datasets for remaining analysis: Dataset 1, which has the outgroup L. watsoni samples included, and Dataset 2, which has only L. littlejohni samples.
Population structure
To investigate population structure, we implemented the Bayesian clustering approach in STRUCTURE on Dataset 1 (Outgroup included) using the admixture model and correlated allele frequency due to short sampling distance between the different river catchments on the Woronora Plateau. We assessed values of K from 4 to 10 with 3 independent runs with 20, 000 burnin and 50 000 MCM iterations for each value of K. Output files from Structure were processed with StructureSelector (Li & Liu 2018) and we used the Puechmaille method to select the best K (Puechmaille 2016). The Puechmaille method employs four estimators (MedMeaK, MaxMeaK, MedMedK and MaxMedK) to determine the best number of clusters (Puechmaille 2016). This method helps to alleviate the issue of uneven sampling (Puechmaille 2016), which is present in the current dataset due to small sample size in the Blue Mountains region. An individual is assigned to a cluster if its arithmetic mean (MedMeak and MaxMeak) or its median (MedMeDK and MaxMedK) membership coefficient to that cluster is greater than the threshold used. To assess the performance of the estimators and prevent over estimation of the true number of clusters we ran replicates of the Puechmaille method using increasing thresholds (0.5, 0.6, 0.7, 0.8). Once best K was selected, we ran STRUCTRE again with the preferred K for 20, 000 burnin and 100, 000 MCMC iterations. A plot identifying the population clusters was created using CLUMPAK through the StructureSelector web interface (Kopelman et al. 2015).
We conducted a genetic test of neutrality by calculating Tajima’s D for all samples and for each individual population in Dataset 2 (Tajima 1989). This test was conducted using the hierfstat V0.5–10 package in R studio and the TajimaD.dosage function after transforming the data into dosage data that reports the number of copies of each allele using the fstat2dos function (Goudet 2005).
Following the test of neutrality, we applied further filtering using the DartR package to Dataset 1 and Dataset 2 to remove loci that were significantly not in Hardy–Weinberg Equilibrium and increased minor allele frequency threshold to 0.02 (RStudio Team 2022; Mijangos et al. 2022). To test for Hardy–Weinberg Equilibrium, we used gl.filter.hwe from the DartR package with individuals grouped by population cluster. This function uses observed frequencies of reference homozygotes, heterozygotes and alternate homozygotes to filter out loci that show departure from HWE. Regarding settings for this function, we employed the exact test as it is recommended in most cases (Wigginton et al. 2005) and used the selome method to estimate p-values with an alpha value of 0.05. The selome method computes p = values as the sum or probabilities of all samples less or equally likely as the current sample (Graffelman 2015). P-values were corrected for multiple tests using the BY method based on Benjamini & Yekutieli (2001), which controls the false discovery rate. The function gl.report.ld.map, from the Dart R package, was employed to test for linkage disequilibrium within populations. This function reports pairwise linkage disequilibrium (LD) for SNPs for which the LD measure is > 0 in all populations and uses a threshold of R2 = 0.20 (Delourme et al. 2013; Li et al. 2014).
Population genetic differentiation
We computed pairwise FST (Weir and Cockerham 1984) using the clusters from the STRUCTURE analysis to further assess the degree of genetic differentiation between population clusters. This was computed using gl.fst.pop from the DartR package and with 1000 bootstrap replications to evaluate whether FST values were significantly different from zero (Gruber et al. 2018; Mijangos et al. 2022).
Isolation by distance and isolation by environment
We conducted a Mantel test of genetic differentiation vs geographic distance between all pairs of L. littlejohni populations, using gl.ibd with 999 permutations from the DartR package on Dataset 2 in R studio (species ingroup only) (Mijangos et al. 2022; RStudio Team 2022). For this test we used Euclidean distance, and we transformed the geographic distances using log(Dgeo + 0.01) as some individuals have identical GPS coordinates that will otherwise result in a geographical distance of 0.
We conducted a partial redundancy analysis in R studio to investigate the influence of environment (Isolation by Environment) and spatial features (Isolation by Distance) on the distribution of genetic variation in Dataset 2 (species ingroup only) (Orsini et al. 2013; Capblanc & Forester 2021: RStudio Team 2022). Partial redundancy analysis cannot handle missing values, therefore, the 0.58% of loci with missing data were substituted with values taken from the nearest neighbouring individual without a missing value using a population-by-population basis. This substitution was conducted using the gl.impute function from the Dart R package. The “neighbour” method as it is recommended for small populations and is nearest neighbour is the individual with the smallest Euclidean distance from the focal individual (Mijangos et al. 2022). The response variable was individual genotypes and we used 3 sets of variables (1) environmental, (2) geography, and (3) genetic structure as explanatory or conditioning variables. To select environmental variables, we downloaded 22 Bioclim variables and elevation data from Atlas of Living Australia Spatial Portal for each individual GPS point (Belbin 2011; Xu & Hutchinson 2011). To determine which environmental variables to include in our partial redundancy analysis we ran a forward selection model using the rda and ordiR2step functions from the R package vegan V2.5.7 to identify variables significantly associated with genetic variation (Oksanen et al. 2013) (Table S2). We then removed environmental variables that were strongly correlated with each other (Pearson’s correlation > 0.60) with preference for those that had higher R2 values and AIC in the forward selected model. The final environmental variables were radiation seasonality, precipitation of driest month, minimum temperature of coldest month, and isothermally. Geographic variables used were the GPS coordinates of individual frogs (Latitude and Longitude). Genetic structure variables were the scores along the first five axes of a genetic Principal Coordinate Analysis conducted on Dataset2 using gl.pcoa from the DartR package (Gruber et al. 2018). We selected the first five axis as they all explained > 1% of variance, totalling to 21.8%. Once variables were finalised, we ran RDAs for a full model (Climate, Geography, Genetic Structure) and one for each set of explanatory variables conditioned by the remaining sets to variables (e.g., Climate conditioned by geography and genetic structure). This allows us to partition the percentage of genetic variance explained by each of the explanatory variables. R squared values were adjusted and an ANOVA with 9999 permutations was run for each model to obtain a p-value.
Genetic diversity
We employed gl.report.heterozygosity from the DartR package using Dataset 2 (species ingroup only) to calculate the observed heterozygosity, expected heterozygosity, unbiased expected heterozygosity, and FIS. Standard deviation for FIS was obtained using boot.ppfis from the hierfstat V0.5–10 package with 100 bootstraps (Goudet 2005).
Mean kinship and inbreeding
Mean kinship is considered one of the best metrics for managing genetic diversity and can help identify the best source populations for augmentation (Frankham et al. 2019). We employed the related V0.8 R package to estimate within population kinship (Pew et al. 2015). There are several relatedness estimators that use the genetic information differently and can yield different estimates (Wang 2011), thus, to determine the best relatedness estimator for our data, we used the function compareestimator from the related package. We implemented this function using a subset of 200 randomly selected loci to compare the seven different relatedness estimators. This function simulates individuals of known relatedness using the loci provided, which then allow users to evaluate the correlation between observed and expected relatedness (Pew et al. 2015). The quellergt estimator, described in Queller and Goodnight (1989) performed the best (Pearson’s r = 0.894), thus used to estimate relatedness. We also employed the lynchrd estimator as it calculates individual inbreeding where as the quellergt estimator does not. This measure of inbreeding (F) is based on the probability of alleles at a random locus being identical by descent (Lynch and Ritland 1999), while FIS indicates the proportion of the variance in a subpopulation contained in the individual. Genetic relatedness among all pairs of L. littlejohni in Dataset 2 were estimated using the quellergt and lynchrd estimators using the function coancestry with 1000 bootstraps and inbreeding allowed. We averaged relatedness to get within population mean kinship and we averaged F for each population.
Effective population size
Effective population sizes were estimated using Dataset 2 and the program NE estimator V (Do et al. 2014). We ran NE Estimator using the Linkage Disequilibrium method with random mating and reported the estimates at a critical allele frequency of 0.05 for each sampling region.
Results
Survey results
Over 185 survey occasions, we caught 265 unique individual frogs with 31 frogs (11.7%) recaptured at various times (Table S1). Recapture rates were the highest in the Central Coast Range with two sites accounting for 48.3% of all recaptures. Twenty sites had no recaptures over the study period. The highest number of individuals caught at a site was 30. No adult frogs were ever found at 2 sites, but tadpoles were detected there.
Population structure
Dataset 1 (outgroup included) had 1,049 loci and 312 individuals post initial filtering. All four of the Puechmaille estimators indicated that the optimal K was six clusters (Fig. 2; Fig. S2). This did not change when using higher threshold values. The six clusters are 1) L. watsoni, 2) Blue Mountains,3) Central Coast Range, 4) O’Hare’s Catchment, 5) Cataract Catchment, and the final cluster is formed by 6) Cordeaux Catchment, Avon Catchment and the Nepean Catchment (referred to as Cordeaux Cluster). Tajima’s D was between zero and one for all populations. The Cordeaux Catchment and Central Coast Range populations had values below 0.50 (Table 3). Post additional filtering, Dataset 2 had 981 loci and 299 individuals.
Structure results for Dataset 1 (outgroup) K = 6 created using StructureSelector web interface CCR- Central Coast Range (maroon), BM—Blue Mountains (green), OH—O’Hare’s Catchment (purple), CAT—Cataract Catchment (orange), AVON—Avon Catchment (blue), CORD—Cordeaux Cluster (blue), NEP—Nepean Catchment (blue), LW—Litoria watsoni (green)
Population genetic differentiation
Pairwise FST values indicate that the Central Coast Range is the most distinctive L. littlejohni population (Table 1). The Central Coast Range population had the highest pairwise FST with all other L. littlejohni populations. Interestingly, the FST value comparing the Central Coast Range and the O’Hare’s catchment indicated that these populations are almost as genetically differentiated as L. watsoni and O’Hare’s. The same pattern was seen for Central coast range and the Cataract cluster. The Blue Mountains is the second most genetically distinctive L. littlejohni population with pairwise FST values all above 0.10. The three populations on the Woronora Plateau had pairwise FST values over 0.05. This value indicates that these are moderately genetically different from each other, however, we detected some movement between population clusters in the structure plot. Three individuals sampled from the O’Hare’s catchment had approximately 50% membership to the Cataract clusters.
Isolation by distance and isolation by adaptation
The Mantel test indicated a significant association between geographical and genetic distances (Mantel’s r = 0.658, r2 = 0.433, p = 0.001). The RDA full model significantly explained 24.1% of the genetic variance across individual frogs (Table 2). The structure model explained 27.1% of the model variance while climate and geography explained 6.4% and 2.6% respectively. Over 63.9% of the variance in the full model was confounded, indicating a strong confounding effect due to collinearity of genetic structure, geography, and climate (Capblanc & Forester 2021).
Genetic diversity, effective population size and kinship
Effective population size was relatively low across all populations with the highest value below 200 in the Cordeaux Cluster (Table 3). The Blue Mountains, Central Coast Range and Cataract populations all had effective population sizes below 50, with the Blue Mountains having the lowest. These smaller populations also had lower observed heterozygosity than those with effective population size over 50 (Cordeaux and O’Hare’s). For all populations, the expected heterozygosity was higher than observed, but this pattern was more pronounced for populations with effective population size below 50. Mean kinship, F, and FIS were highest in the Central Coast Range, Blue Mountains, and Cataract catchment.
Discussion
Obtaining estimates of population size and connectivity is a critical step within conservation research, however, for many threatened amphibians this is a difficult task due to the cryptic nature or rarity of such species. In this study of L. littlejohni, where we were unable to obtain sufficient recaptures to estimate population sizes through traditional mark-recapture methods, however, we were able to estimates effective population size in five genetically distinct populations using genetic methods. Additionally, we revealed the restriction of gene flow at the regional and local scale. This additional knowledge would be unlikely obtained through standard capture-recapture methods. Two population clusters are clearly geographically and genetically isolated, but the other three are located on the same plateau. Climate and geography were not strong drivers of genetic isolation. High levels of inbreeding, small population size and reduced genetic diversity were detected in the two geographically isolated populations and one of the populations on the Woronora plateau. These results provide strong evidence that the rarity of this frog is likely due to small population sizes rather than low detection rates and urgent conservation action is needed to ensure the persistence of this threatened frog.
Population structure
When comparing the major sampling regions, strong genetic isolation was observed in L. littlejohni. Based on the results of the pRDAs, we suggest that genetic drift due to population contractions is the main driver of genetic variation across populations, rather than large climatic difference or geographic distance between populations (Capblancq & Forester 2021; Coleman et al. 2013). While Isolation by Distance was significant, the partial RDA indicated that only a small percentage of genetic variation could be attributed to geography. In the partial RDA models, population genetic structure was the strongest predictor of genetic variation indicating that demographic history rather isolation by adaption or isolation by distance have driven patterns of genetic variation in L. littlejohni (Capblancq & Forester 2021). However, these variables were strongly confounded and over 63% of variation in the full model could not be exclusively attributed to climate, geography or genetic structure alone. This suggests any future work to investigate loci under selection or conduct further landscape genetic analysis in L. littlejohni must consider ways to account for this collinearity (Frichot et al. 2015; Hoban et al 2016; Capblancq & Forester 2021). The test of neutrality further supports the notion of genetic drift following population crash as the best explanation of modern population differentiation in L. littlejohni. All populations had positive Tajima’s D values, which may indicate very balancing selection or sudden population contraction (Tajima 1989). However, all values had quite low absolute value indicating that any selection would be weak.
Increased genetic structuring of populations has been linked to disease-related population contractions in numerous vertebrate species. In these species, population contractions led to increased population fragmentation and exacerbated genetic drift causing populations to become more genetically differentiated (Phillips et al. 2020; Serieys et al. 2015; Lachish et al. 2011). Both the Central Coast Range and Blue Mountains have effective population sizes below 50, high inbreeding values and lower genetic diversity compared to other L. littlejohni populations, indicating that these populations have indeed suffered from population bottlenecks. Between these two regions are largely continuous natural areas consisting of seemingly suitable habitat where there were historical sightings of L. littlejohni, thus these regions were once connected (Mahony et al. 2020). Due to a lack of pre-chytrid genetic samples, we cannot confirm chytrid as the driver of L. littlejohni declines in this study, although this disease is the most likely driver of sudden decline in this winter active, stream and pond frog (Hero 2005). However, it seems likely that these two populations have become further isolated both geographically and genetically due to disease driven population declines with the resulting small population sizes exacerbating the consequences genetic drift.
Large scale damming and population crashes due to disease may explain the strong genetic isolation between the Blue Mountains and Woronora Plateau populations. Damming of riverine systems alters population connectivity by fragmenting suitable habitat (Heggenes & Roed 2006; Emel & Storfer 2012; Hansen et al. 2014; Werth et al. 2014). In other frog species, river regulation has led to changes in frog species composition, increased genetic isolation, and loss of genetic diversity (Naniwadekar & Vasudevan 2014; Peek et al. 2021, Grummer & Leache 2017). We hypothesize that the construction of the Warragamba Dam in the 1960s may have disrupted gene flow between the Blue Mountains and Woronora populations. The Warragamba dam creates Lake Burragorang, however, L. littlejohni does not occupy large lakes. Rather, this frog is typically associated with first order streams and small fire dams (Mahony et al. 2020; Lemckert 2010). Thus, the lake may act as barrier due to unsuitable habitat.
The degree of genetic structuring observed on the Woronora Plateau was unexpected as the landform is contiguous and there is little urban development beyond major roads. This region has the largest population of L. littlejohni, thus it was anticipated that gene flow would occur between the sampled river catchments. Instead, we detected three genetic clusters with low to moderate genetic differentiation (O’Hares, Cataract and Cordeaux; Fig. 2). Distinct genetic structure at a local scale is typically seen in species that have high site fidelity and inherently low dispersal abilities (Zickovich & Bohonak 2007; Richardson et al. 2021), however, we do not think this is the case for L. littlejohni. The geographic size of the Cordeaux cluster (> 84km2) indicates that L. littlejohni is not limited by its ability to disperse across substantial distances. We propose three factors that may explain the fine-scale structuring of L. littlejohni populations: 1) roads that prevent movement between catchments; 2) dispersal is restricted to creek lines; and 3) habitat disturbance that has isolated populations locally. As discussed with the other populations, the genetic structuring may also be exacerbated by general distribution-wide declines due to chytrid.
Roads (1) restrict movement of a variety of vertebrate species, leads to genetic isolation of populations (Hale et al. 2013; Holderegger & Di Giulio 2010; Serieys et al. 2015; Youngquist et al. 2017). For example, major urban infrastructure (housing and dual carriage roads) has been linked to the genetic sub-division of southern bell frog (L. raniformis) (Hale et al. 2013), and the presence of highways is predictive of genetic distance in Blanchard’s cricket frogs (Acris blanchardi) (Youngquist et al. 2017). The Woronora Plateau has several large dual carriageway roads which traverse it (Picton Road and Appin Road) (Fig. 1), which are possible barriers to dispersal in L. littlejohni.
Dispersal primarily along creek lines (2) rather than through terrestrial landscapes may contribute to isolation of populations. For some aquatic and semi aquatic species, patterns of gene flow reflect the hierarchical nature of river systems and/or the isolation of populations to independent river basins or drainages (Brauer et al. 2016; Coa et al. 2020; Fonseca et al. 2021). Whilst the river catchments on the Woronora Plateau are geographically close to each other, they are parts of different major river drainages (Fig. 1). The Cordeaux Cluster and Cataract Catchment flow into the Hawkesbury-Nepean River system, while the O’Hare’s Catchment flows into the Georges River/Tucoerah River. O’Hare’s and the Cataract site is essentially the border of these two major river catchments. Although we did detect movement between Cataract and O’Hare’s sites, the moderate genetic differentiation of these two clusters indicates movement is not common despite the short distance between them (less than 5 km between Cataract and closest O’Hare’s site). As L. littlejohni tadpoles are aquatic, larval dispersal along creeks (particularly during high rain periods) is highly probable. Additionally, during our surveys we often encountered adult frogs utilizing the sandstone bedrock of creek lines to move between water bodies. We only detected a frog more than thirty metres from water once, despite accessing sites by walking. There have been reports of L. littlejohni found under rocks away from streams during other fauna surveys (G. Daly unpubl. data), thus some terrestrial movement does occur, but it is suspected to be limited.
In addition to roads, the main habitat disturbance (3) occurring in the Woronora is longwall coal mining. Longwall mining can lead to the diversion of water underground due to subsidence causing cracks in the surface rock (Booth 2006). The Cataract River Catchment has been undermined extensively, however, to what degree this mining has impacted frog species is unclear due to a lack of research (WaterNSW 2016). In the last ten years, sightings of L. littlejohni within Cataract have been limited in number and are restricted to small pockets within the catchment. It is plausible that mining activity has fragmented populations locally as drying of creeks may result in limited successful dispersal events. However, longwall mining also occurs within the Cordeaux Cluster, which has no population sub-structuring, thus there is no clear trend as to how creek drying due to mining impacts frog movement.
Small populations and inbreeding
The small effective population size across all L. littlejohni populations indicate that the rarity of this species is indeed due to historical declines and not just strict habitat preference or cryptic behaviour. These estimates are also of conservation concern as the fate of fragmented populations is determined by the effective population size of the isolated population, rather than the species as a whole (Frankham et al. 2019). While the effective population sizes only reflect that of the sampled areas, as we sampled a large proportion of known currently occupied sites, thus it is unlikely that these estimates would change dramatically with additional samples. The three smallest populations (Central Coast range, Blue Mountains, and Cataract) also have higher inbreeding coefficients for both F and FIS, lower observed heterozygosity and higher mean kinship than the O’Hare’s and Cordeaux clusters. Together these factors put populations at risk of reduced adaptive potential, inbreeding depression, and increased sensitivity to stochastic events (Frankham et al. 2019). Additionally, populations may be impacted by mutational meltdown, a type of extinction vortex (Keller & Waller 2002). Mutational meltdown is a process in which genetic and demographics processes mutually reinforce one another as deleterious mutations become fixed and accumulate within the population. These mutations lead to further reductions in population size and in turn increases the rate that mutations are accumulated and the speed at which populations decline in size (Lynch & Gabriel 1990). Alternatively, deleterious alleles may be purged from populations with high inbreeding and small population size. Inbreeding is expected to increase the expression of deleterious alleles as more individuals will be homozygotes for recessive alleles. Natural selection should select against such individuals in the population, leading to less individuals overall with the recessive deleterious alleles. However, the extent of genetic purging depends on many genetic factors and the environment in which it takes place (Keller & Waller 2002).
The coefficient of inbreeding recommended by Frankham et al. (2019), F, uses identity by descent methods to estimate cumulative inbreeding over generations (Frankham et al. 2019). On the other hand, FIS is used more widely in population genetic research, but only detects deviations from random mating in recent generations. In the present study, we have populations in three different states (1) High F and high FIS, (2) Low F and low FIS, and (3) High F but low FIS. The Cordeaux cluster is in State 1 (Low F and FIS) and has the largest population size, indicating that this population may not have faced large population declines or the population maintained a large enough size to prevent changes in random mating patterns. The Central Coast Range, Blue Mountains and Cataract are all in State 2 (High F and FIS) and have small population sizes, indicating that these populations have declined and not recovered. The final population, O’Hare’s, is in State 3 (High F, but low FIS) and has a small but larger population than those in State 2. The difference in inbreeding values may indicate that the current population is not deviating from random mating, but that in the past the population was small enough for inbreeding to be present. Thus, O’Hare’s likely experienced decline and has recovered to some extent. There are several examples of chytrid susceptible species that have declined in some parts of their range but persist or have recovered in others (Osborne et al. 1996; Retallick et al. 2004; Wassens 2008; Hamer et al 2010; Mahony et al 2013; Newell et al. 2013; Lips 2016). Habitat quality, changes to disease dynamics, recruitment rates and population connectivity have all been linked to stabilizations and recoveries of frog species (Scheele et al. 2014; McDonald et al. 2005; Scheele et al. 2017; Mc Knight et al. 2019). As L. littlejohni populations appear to differ in recovery history they may provide a useful model for further investigation into drivers of recovery.
Management recommendations
Through the application of genetic methods, conservation managers will be able to make decisions for L. littlejohni with more clarity going forward. The small effective populations, high inbreeding, reduced genetic diversity and high mean kinship detected in some L. littlejohni population indicates that urgent on-ground action is needed to increase population sizes. Determining habitat preferences and protecting vital breeding habitat will be important for supporting successful recruitment within populations. Recommended research includes assessing whether inbreeding depression is present in populations with high FIS and F. Inbreeding depression in other frogs has been linked to reduced sperm quality and reduced offspring survival (Hinkson & Poo 2020; Anderson et al. 2004), which obviously has implications for population viability of threatened species. Additionally, we recommend assessing whether assisted gene flow will aid in genetic rescue or lead to outbreeding depression (Frankham et al. 2019). The Cordeaux and O’Hare’s clusters are recommended as potential source populations as they contain the highest genetic diversity, lowest inbreeding, and largest populations. Additionally, the metrics for coefficient of inbreeding and mean kinship have both been used in other species to assess or plan translocations (Cowen et al. 2021; Farquharson et al. 2021), thus the values reported here provide a baseline for future conservation management.
Global significance/conclusions
This study demonstrates the power of pairing genetic methods with traditional surveys to study rare and difficult to track species. Without employing genetic methods in the present study, we could have not resolved the issue of low sampling rates and low recapture rates that prevented successful mark-recapture methods. Additionally, the SNP data provided insight into potential inbreeding depression, which highlights an urgent need for conservation actions and further research, that would not have been detected through visual encounters surveys alone. Incorporating genetics into conservation research can have a profound impact on both our understanding and our ability to provide adequate management of threatened species.
Data availability
The SNP databases generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Allendorf FW, Hohenlohe PA, Luikart G (2010) Genomics and the future of conservation genetics. Nat Rev Genet 11:697–709
Alton LA, Franklin CE (2017) Drivers of amphibian declines: effects of ultraviolet radiation and interactions with other environmental factors. Clim Chang Responses 4:6. https://doi.org/10.1186/s40665-017-0034-7
Amato G, DeSalle R, Ryder OA, Rosenbaum HC (2009) Conservation genetics in the age of genomics. Columbia University Press, New York
Andersen LW, Fog K, Damgaard C (2004) Habitat fragmentation causes bottlenecks and inbreeding in the European tree frog (Hyla arborea). The Royal Society 271:1293–1302
Anderson EC, Dunham KK (2008) The influence of family groups on inferences made with the program Structure. Mol Ecol Resour 8:1219–1229. https://doi.org/10.1111/j.1755-0998.2008.02355.x
Belbin, Lee (2011) The Atlas of Livings Australia’s Spatial Portal. In, Proceedings of the Environmental Information Management Conference 39–43 Santa Barbara
Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29:1165–1188
Berger L, Speare R, Daszak P, Green DE, Cunningham AA, Goggin CL, Parkes H (1998) Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci 95:9031–9036. https://doi.org/10.1073/pnas.95.15.9031
Booth CJ (2006) Groundwater as an environmental constraint of longwall coal mining. Environ Geol 349(6):796–803
Bower DS, Lips KR, Schwarzkopf L, Georges A, Clulow S (2017) Amphibians on the brink. Science 357:454–455
Bradburd GS, Ralph PL, Coop GM (2013) Disentangling the effects of geographic and ecological isolation on genetic differentiation. Evolution 67:3258–3273. https://doi.org/10.1111/evo.12193
Bradford T, Pickett M, Donnellan S, Gardner MG, Schofield J (2018) Conservation genomics of an endangered subspecies of southern emu-wren stipiturus malachurus (Passeriformes: Maluridae). Emu - Austral Ornithology 118:258–268. https://doi.org/10.1080/01584197.2017.1422391
Brauer CJ, Hammer MP, Beheregaray LB (2016) Riverscape genomics of a threatened fish across a hydroclimatically heterogeneous river basin. Mol Ecol 20:5093–5113. https://doi.org/10.1111/mec.13830
Cao R, Somaweera R, Brittain K, FitzSimmons NN, Georges A, Gongora J (2020) Genetic structure and diversity of Australian freshwater crocodiles (Crocodylus johnstoni) from the Kimberley Western Australia. Conserv Genet 21:421–429. https://doi.org/10.1007/s10592-020-01259-5
Capblancq T, Forester BR (2021) Redundancy analysis: a Swiss Army Knife for landscape genomics. Methods Ecol Evol 12:2298–2309. https://doi.org/10.1111/2041-210X.13722
Coleman RA, Weeks AR, Hoffmann AA (2013) Balancing genetic uniqueness and genetic variation in determining conservation and translocation strategies: a comprehensive case study of threatened dwarf galaxias, Galaxiella pusilla (Mack) (Pisces: Galaxiidae). Mol Ecol 22:1820–1835. https://doi.org/10.1111/mec.12227
Cowen SJ, Comer S, Wetherall JD, Groth DM (2021) Translocations and their effect on population genetics in an endangered and cryptic songbird the Noisy Scrub-bird Atrichornis clamosus. Emu - Austral Ornithology 121:33–44. https://doi.org/10.1080/01584197.2021.1888125
Cushman SA (2006) Effects of habitat loss and fragmentation on amphibians: a review and prospectus. Biol Cons 128:231–240. https://doi.org/10.1016/j.biocon.2005.09.031
Delourme R, Falentin C, Fomeju BF et al (2013) High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napusL. BMC Genomics 14(1):120. https://doi.org/10.1186/1471-2164-14-120
Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR (2014) NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour 1:209–214. https://doi.org/10.1111/1755-0998.12157
Emel SL, Storfer A (2012) A decade of amphibian population genetic studies: synthesis and recommendations. Conserv Genet 13:1685–1689. https://doi.org/10.1007/s10592-012-0407-1
Farquharson KA, McLennan EA, Wayne A, Smith M, Peel E, Belov K, Hogg CJ (2021) Metapopulation management of a critically endangered marsupial in the age of genomics. Global Ecol Conserv 31:e01869. https://doi.org/10.1016/j.gecco.2021.e01869
Fonseca EM, Garda AA, Oliveira EF et al (2021) The riverine thruway hypothesis: rivers as a key mediator of gene flow for the aquatic paradoxical frog Pseudis tocantins (Anura, Hylidae). Landscape Ecol 36:3049–3060. https://doi.org/10.1007/s10980-021-01257-z
Frankham R (2010) Challenges and opportunities of genetic approaches to biological conservation. Biol Cons 143:1919–1927. https://doi.org/10.1016/j.biocon.2010.05.011
Frankham R, Ballou JD, Ralls K, Eldridge M, Dudash MR, Fenster CB, Sunnucks P (2019) A practical guide for genetic management of fragmented animal and plant populations. Oxford University Press, Oxford
Frichot E, Schoville SD, De Villemereuil P, Gaggiotti OE, François O (2015) Detecting adaptive evolution based on association with ecological gradients: orientation matters! Heredity 115:22–28. https://doi.org/10.1038/hdy.20
Georges A, Gruber B, Pauly GB, White D, Adams M, Young MJ, Unmack PJ (2018) Genomewide SNP markers breathe new life into phylogeography and species delimitation for the problematic short-necked turtles (Chelidae: Emydura) of eastern Australia. Mol Ecol 27:5195–5213. https://doi.org/10.1111/mec.14925
Gill L, Klop-Toker K, Hayward MW (2021) Identifying the breeding activity and phenology of the Northern and Southern Heath Frog through sound analysis. Honours thesis, University of Newcastle, Newcastle, Australia
Gillespie GR, McNabb E, Gaborov R (2016) The biology and status of the large brown tree frog Litoria littlejohni in Victoria. Vic Nat 133:128–138
Goudet J (2005) HIERFSTAT a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186. https://doi.org/10.1111/j.1471-8286.2004.00828.x
Goudet J, Kay T, Weir BS (2018) How to estimate kinship. Mol Ecol 27(20):4121–4135. https://doi.org/10.1111/mec.14833
Graffelman J (2015) Exploring diallelic genetic markers: the Hardy Weinberg package. J Stat Softw 64:1–23
Gruber B, Unmack PJ, Berry OF, Georges A (2018) dartr: An r package to facilitate analysis of SNP data generated from reduced representation genome sequencing. Mol Ecol Resour 3:691–699. https://doi.org/10.1111/1755-0998.12745
Grummer JA, Leaché AD (2017) Do dams also stop frogs? Assessing population connectivity of coastal tailed frogs (Ascaphus truei) in the North Cascades National Park Service Complex. Conserv Genet 18:439–451. https://doi.org/10.1007/s10592-016-0919-1
Hale JM, Heard GW, Smith KL, Parris KM, Austin JJ, Kearney M, Melville J (2013) Structure and fragmentation of growling grass frog metapopulations. Conserv Genet 14:313–322. https://doi.org/10.1007/s10592-012-0428-9
Hamer AJ, Lane SJ, Mahony MJ (2010) Using probabilistic models to investigate the disappearance of a widespread frog-species complex in high-altitude regions of south-eastern Australia. Animal Conserv 13:275–285. https://doi.org/10.1111/j.1469-1795.2009.00335.x
Hansen MM, Limborg MT, Ferchaud A-L, Pujolar J-M (2014) The effects of Medieval dams on genetic divergence and demographic history in brown trout populations. BMC Evol Biol 14:122. https://doi.org/10.1186/1471-2148-14-122
Harrisson KA, Magrath MJL, Yen JDL et al (2019) Lifetime fitness costs of inbreeding and being inbred in a critically Endangered Bird. Curr Biol 16:2711-2717.e4. https://doi.org/10.1016/j.cub.2019.06.064
Hayes TB, Khoury V, Narayan A, Nazir M, Park A, Brown T, Stueve T (2010) Atrazine induces complete feminization and chemical castration in male African clawed frogs (Xenopus laevis). Proc Natl Acad Sci 107:4612–4617. https://doi.org/10.1073/pnas.0909519107
Heggenes J, Røed KH (2006) Do dams increase genetic diversity in brown trout (Salmo trutta)? Microgeographic differentiation in a fragmented river. Ecol Freshw Fish 15:366–375. https://doi.org/10.1111/j.1600-0633.2006.00146.x
Hero JM, Williams SE, Magnusson WE (2005) Ecological traits of declining amphibians in upland areas of eastern Australia. J Zool 267:221–232
Hinkson KM, Poo S (2020) Inbreeding depression in sperm quality in a critically endangered amphibian. Zoo Boil 391:197–204. https://doi.org/10.1002/zoo.21538
Hoban S et al (2016) Finding the genomic basis of local adaptation: Pitfalls, practical solutions, and future directions. Am Nat 188:379–397
Hohenlohe PA, Funk WC, Rajora OP (2021) Population genomics for wildlife conservation and management. Mol Ecol 30:62–82. https://doi.org/10.1111/mec.15720
Holderegger R, Di Giulio M (2010) The genetic effects of roads: a review of empirical evidence. Basic Appl Ecol 11:522–531
IUCN (2012) Guidelines for application of IUCN red list criteria at regional and national levels: version 4.0.
Kamvar ZN, Tabima JF, Grünwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281
Kamvar ZN, Brooks JC, Grünwald NJ (2015) Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front Genet 6:208. https://doi.org/10.3389/fgene.2015.00208
Kats LB, Ferrer RP (2003) Alien predators and amphibian declines: review of two decades of science and the transition to conservation. Divers Distrib 9:99–110. https://doi.org/10.1046/j.1472-4642.2003.00013.x
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241. https://doi.org/10.1016/S0169-5347(02)02489-8
Klop-Toker K, Wallace S, Stock S, Hayward MW, Mahony M (2021) The Decline of Australian Heath Frogs and summary of current threats. Imperilled. https://doi.org/10.1016/B978-0-12-821139-7.00145-8
Kopelman NM, Mayzel J, Mattias J, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across. K Mol Ecol Res 15(5):1179–1191. https://doi.org/10.1111/1755-0998.12387
Lachish S, Miller KJ, Storfer A, Goldizen AW, Jones ME (2011) Evidence that disease-induced population decline changes genetic structure and alters dispersal patterns in the Tasmanian devil. Heredity 106:172–182. https://doi.org/10.1038/hdy.2010.17
Laurance WF (2008) Global warming and amphibian extinctions in eastern Australia. Austral Ecol 33:1–9. https://doi.org/10.1111/j.1523-1739.2007.00777.x
Lemckert F (2004) The biology and conservation status of the heath frog Litoria littlejohni. Herpetofauna 34:99–104
Lemckert F (2010) Habitat relationships and presence of the threatened heath frog Litoria littlejohni (Anura: Hylidae) in central New South Wales. Australia Endanger Species Res 11:271–278
Li Y-L, Liu J-X (2018) StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol Ecol Resour. 18:176–177. https://doi.org/10.1111/1755-0998.12719
Li X, Han Y, Wei Y, Acharya A, Farmer AD et al (2014) Development of an alfalfa SNP array and its use to evaluate patterns of population structure and linkage disequilibrium. PLoS One 9(1):e84329. https://doi.org/10.1371/journal.pone.0084329
Lip K (2016) Overview of chytrid emergence and impacts on amphibians. Phil Trans r Soc B 371:20150465. https://doi.org/10.1098/rstb.2015.0465
Lynch M, Gabriel W (1990) Mutation load and the survival of small populations. Evolution 44:1725–1737. https://doi.org/10.1111/j.1558-5646.1990.tb05244.x
Lynch M, Ritland K (1999) Estimation of pairwise relatedness with molecular markers. Genetics 152(4):1753–1766. https://doi.org/10.1093/genetics/152.4.1753
Mahony MJ, Hamer AJ, Pickett EJ, McKenzie DJ, Stockwell MP, Garnham JI, Keely CC, Deboo ML, O’Meara J, Pollard CJ, Clulow S, Lemckert FL, Bower DS, Clulow J (2013) Identifying conservation and research priorities in the face of uncertainty: a review of the threatened bell frog complex in eastern Australia. Herpetol Conserv Biol 8(3):519–538
Mahony M, Moses B, Mahony SV, Lemckert FL, Donnellan S (2020) A new species of frog in the Litoria ewingii species group (Anura: Pelodryadidae) from south-eastern Australia. Zootaxa 4858:201–230. https://doi.org/10.11646/zootaxa.4858.2.3
McCartney-Melstad E, Shaffer HB (2015) Amphibian molecular ecology and how it has informed conservation. Mol Ecol 20:5084–5109
McDonald KR, Méndez D, Müller R, Freeman AB, Speare R (2005) Decline in the prevalence of chytridiomycosis in frog populations in North Queensland, Australia. Pacific Conserv Biol 11:114–120. https://doi.org/10.1071/PC050114
McKnight DT, Lal MM, Bower DS, Schwarzkopf L, Alford RA, Zenger KR (2019) The return of the frogs: the importance of habitat refugia in maintaining diversity during a disease outbreak. Mol Ecol 28:2731–2745. https://doi.org/10.1111/mec.15108
Mendelson JR et al (2006) Confronting amphibian declines and extinctions. Science 313:48
Mijangos J, Gruber B, Berry O, Pacioni C, Georges A (2022) dartR v2: An accessible genetic analysis platform for conservation, ecology and agriculture. Methods Ecol Evol. https://doi.org/10.1111/2041-210X.13918
Miller JM, Quinzin MC, Edwards DL, Eaton DAR, Jensen EL, Russello MA, Gibbs JP, Tapia W, Rueda D, Caccone A (2018) Genome-wide assessment of diversity and divergence among Extant Galapagos Giant Tortoise Species. J Heredity 109(6):611–619. https://doi.org/10.1093/jhered/esy031
Naniwadekar R, Vasudevan K (2014) Impact of Dams on Riparian Frog communities in the Southern Western Ghats. India Divers 6(3):567–578. https://doi.org/10.3390/d6030567
Newell DA, Goldingay RL, Brooks LO (2013) Population Recovery following Decline in an Endangered Stream-Breeding Frog (Mixophyes fleayi) from Subtropical Australia. PLoS One 8(3):e5855. https://doi.org/10.1371/journal.pone.0058559
O’Connell KA, Mulder KP, Maldonado J, Currie KL, Ferraro DM (2019) Sampling related individuals within ponds biases estimates of population structure in a pond-breeding amphibian. Ecol Evol 9:3620–3636. https://doi.org/10.1002/ece3.4994
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara RB, Wagner H (2013) Package ‘vegan’, Community ecology package version 2:1-295, https://cran.r-project.org/web/packages/vegan
Orsini L, Vanoverbeke J, Swillen I, Mergeay J, De Meester L (2013) Drivers of population genetic differentiation in the wild: isolation by dispersal limitation isolation by adaptation and isolation by colonization. Mol Ecol 22:5983–5999. https://doi.org/10.1111/mec.12561
Osborne WS, Littlejohn MJ, Thomson SA (1996) Former distribution and apparent disappearance of the Litoria aurea complex from the Southern Tablelands of New South Wales and the Australian Capital Territory. Australian Zool 30(2):190–198. https://doi.org/10.7882/AZ.1996.011
Patterson N, Price AL, Reich D (2006) Population structure and eigenanalysis. PLoS Genet 2:e190. https://doi.org/10.1371/journal.pgen.0020190
Peek RA, O’Rourke SM, Miller MR (2021) Flow modification associated with reduced genetic health of a river-breeding frog Rana boylii. Ecosphere 12(5):e03496. https://doi.org/10.1002/ecs2.3496
Pew J, Muir PH, Wang J, Frasier TR (2015) related: an R package for analysing pairwise relatedness from codominant molecular markers. Mol Ecol Resour 15:557–561. https://doi.org/10.1111/1755-0998.12323
Phillips P, Livieri TM, Swanson BJ (2020) Genetic signature of disease epizootic and reintroduction history in an endangered carnivore. J Mammal 101:779–789
Puechmaille SJ (2016) The program structure does not reliably recover the correct population structure when sampling is uneven: subsampling and new estimators alleviate the problem. Mol Ecol Resour 16:608–627. https://doi.org/10.1111/1755-0998.12512
Queller DC, Goodnight KF (1989) Estimating relatedness using molecular markers. Evolution 43:258–275
Retallick RWR, McCallum H, Speare R (2004) Endemic Infection of the Amphibian Chytrid Fungus in a Frog Community Post-Decline. PLoS Biol 2(11):e351. https://doi.org/10.1371/journal.pbio.0020351
Richardson JL, Michaelides S, Combs M, Djan M, Bisch L, Barrett K, McGreevy TJ (2021) Dispersal ability predicts spatial genetic structure in native mammals persisting across an urbanization gradient. Evol Appl 14:163–177. https://doi.org/10.1111/eva.13133
RStudio Team (2022). RStudio: Integrated Development for R. RStudio, PBC, Boston, MA URL http://www.rstudio.com/
Scheele BC, Guarino F, Osborne W, Hunter DA, Skerratt LF, Driscoll DA (2014) Decline and re-expansion of an amphibian with high prevalence of chytrid fungus. Biol Conserv 170:86–91. https://doi.org/10.1016/j.biocon.2013.12.034
Scheele BC, Skerratt LF, Grogan LF, Hunter DA, Clemann N, McFadden M, Newell D, Hoskin JC, Gillespie GR, Heard GW, Brannelly L, Roberts AA, Berger L (2017) After the epidemic: Ongoing declines, stabilizations and recoveries in amphibians afflicted by chytridiomycosis. Biol Cons 206:37–46. https://doi.org/10.1016/j.biocon.2016.12.010
Scheele BC et al (2019) Amphibian fungal panzootic causes catastrophic and ongoing loss of biodiversity. Science 363:1459–1463
Serieys LEK, Lea A, Pollinger JP, Riley SPD, Wayne RK (2015) Disease and freeways drive genetic change in urban bobcat populations. Evol Appl 8:75–92
Sexton JP, Hangartner SB, Hoffmann AA (2014) Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution 68:1–15. https://doi.org/10.1111/evo.12258
Skerratt LF, Berger L, Speare R, Cashins S, McDonald KR, Phillott AD, Kenyon N (2007) Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. EcoHealth 4:125–134. https://doi.org/10.1007/s10393-007-0093-5
Speed D, Balding DJ (2015) Relatedness in the post-genomic era: is it still useful? Nat Rev Genet 16(1):33–44
Stuart SN, Chanson JS, Cox NA, Young BE, Rodrigues ASL, Fischman DL, Waller RW (2004) Status and trends of amphibian declines and extinctions worldwide. Science 306:178–1786
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123(3):585–595. https://doi.org/10.1093/genetics/123.3.585
Thackway R, Cresswell ID (1995) An interim biogeographic regionalisation for Australia: a framework for establishing the national system of reserves, Version 4.0. Australian Nature Conservation Agency, Canberra
Wang J (2011) coancestry: a program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol Ecol Resour 11:141–145. https://doi.org/10.1111/j.1755-0998.2010.02885.x
Wang IJ, Bradburd GS (2014) Isolation by environment. Mol Ecol 23:5649–5662. https://doi.org/10.1111/mec.12938
Wang IJ, Summers K (2010) Genetic structure is correlated with phenotypic divergence rather than geographic isolation in the highly polymorphic strawberry poison-dart frog. Mol Ecol 19:447–458. https://doi.org/10.1111/j.1365-294X.2009.04465.x
Wang IJ, Savage WK, Bradley Shaffer H (2009) Landscape genetics and least-cost path analysis reveal unexpected dispersal routes in the California tiger salamander (Ambystoma californiense). Mol Ecol 18:1365–1374. https://doi.org/10.1111/j.1365-294X.2009.04122.x
Wassens S (2008) Review of the past distribution and decline of the Southern Bell Frog Litoria raniformis in New South Wales. Australian Zool 34(3):446–452. https://doi.org/10.7882/AZ.2008.022
WaterNSW, Literature Review of Underground Mining Beneath Catchments and Water Bodies 2016
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370. https://doi.org/10.2307/2408641
Werth S, Schödl M, Scheidegger C (2014) Dams and canyons disrupt gene flow among populations of a threatened riparian plant. Freshw Ecol 59:2502–2515. https://doi.org/10.1111/fwb.12449
Wigginton JE, Cutler DJ, Abecasis GR (2005) A note on exact tests of hardy-weinberg equilibrium. Amer J Human Genet 76(5):887–893. https://doi.org/10.1086/429864
Xu T, Hutchinson MF (2011) ANUCLIM Version 6.1 User Guide. Fenner School of Environment and Society, Australian National University. 90 p. fennerschool.anu.edu.au/files/anuclim61.pdf (Accessed 15 April 2018)
Youngquist MB, Inoue K, Berg DJ, Boone MD (2017) Effects of land use on population presence and genetic structure of an amphibian in an agricultural landscape. Landsc Ecol 32:147–162. https://doi.org/10.1007/s10980-016-0438-y
Zickovich JM, Bohonak AJ (2007) Dispersal ability and genetic structure in aquatic invertebrates: a comparative study in southern California streams and reservoirs. Freshw Biol 52:1982–1996. https://doi.org/10.1111/j.1365-2427.2007.01822.x
Acknowledgements
We acknowledge the Awabakal people as the traditional owners on the lands upon which this study was conceived, and the Tharawal, Awabakal, and Darkinjung people where field work occurred, and we pay our respects to elders past, present and emerging. This work benefitted from the guidance of J. Dawson. We thank D. McKnight, C. Pacioni, and W. Stock for statistical advice and all the volunteers that have helped with sample collection. Fieldwork was undertaken under Scientific Licences from the NSW Office of Environment and Heritage (National Parks and Wildlife), and under Animal Ethics approval from the University of Newcastle Animal Ethics Committee. Fieldwork in NSW State Forests was conducted under a Special Purpose Permit and access to Special Areas was provided through WaterNSW. This work was funded by the Federal Department of Agriculture, Water and Environment and the NSW Government through a partnership between the Saving our Species program and the Environmental Trust.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This project was funded by NSW Government through a partnership between the Saving our Species program and the Environmental Trust and the Federal Department of Agriculture, Water and Environment.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation and data collection were performed by SES, KK-T, SW, OK, RS, SM, and AC. Analysis was performed and the first draft manuscript was written by performed by SE Stock. All authors commented on previous versions of the manuscript and all authors read and approved the final manuscript. Funding was obtained by AC, KK-T, MW Hayward, and MM.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
Animal ethics approval came from the University of Newcastle (A-2019–924 and A2018-816) and work was conducted under scientific licence (SL 102324, SL100190, and SL102105).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Stock, S.E., Klop-Toker, K., Wallace, S. et al. Uncovering inbreeding, small populations, and strong genetic isolation in an Australian threatened frog, Litoria littlejohni. Conserv Genet 24, 575–588 (2023). https://doi.org/10.1007/s10592-023-01522-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-023-01522-5