Abstract
In the current negotiations regarding revision of the Convention on Biological Diversity (CBD) proposals have been made to strengthen the genetic goals, indicators, and targets for wild species in natural habitats by specifying “tolerable” losses of genetic diversity. However, they have not been subjected to evaluations of their continued use over 100 years, a common conservation time frame. I evaluated six scenarios (3, 5 or 10% loss of genetic diversity [heterozygosity] over 8 or 32 years) proposed as targets for revision of genetic indicators in CBD by predicting their consequences on genetic diversity, inbreeding, fitness, and evolutionary potential when applied at the same rate for 100 years. All proposals lead to substantial genetic harm to species when continued for 100 years that will compromise species persistence, especially in the context of environmental change. Consequently, none of the proposals are suitable for inclusion in the CBD. However, alternative indicators are proposed that would reflect improvements in the genetic status of populations and species, namely (1) the number of species and their populations being maintained at sizes sufficient to retain evolutionary potential in perpetuity, and (2) the number of species for which population genetic connectivity has been improved.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Convention on Biological Diversity (CBD) is the first global agreement to cover all aspects of biodiversity. It was drafted in Rio de Janeiro, Brazil in 1992 and became effective on 29th December 1993. This multilateral treaty has three main goals: the conservation of biodiversity, the sustainable use of its components, and the equitable sharing of benefits arising from genetic resources. Here, I am concerned with its role in biodiversity conservation, especially wild species of animals, plants, and fungi in natural habitats.
While the original CBD mentioned genetics, its main focus was on conserving genetic diversity for domestic plants and animals, with wild animals and plants being neglected. A new post-2020 Global Biodiversity Framework, is being developed to guide action through 2030, albeit with delays due to the covid pandemic. There is a strong push to strengthen the genetic content of the convention to include wild animals, plants, and fungi (Hoban et al. 2020; Laikre et al. 2020) and to set goals, indicators, and targets for 2030 and 2050, but there has been little agreement about them. Consequently, an IUCN taskforce was set up in December 2021 to evaluate the various proposals and to report back to the wider committee and I joined it as a representative of the Conservation Planning Specialist Group of IUCN. This material was first prepared as a position paper for that taskforce. As of May 2022 the genetic content of the revised CBD is still being negotiated.
Evaluation of proposed CBD targets for retaining genetic diversity
The targets for retaining genetic diversity (GD) proposed for insertion into the revised CBD appear to have originated from the goal for captive populations, namely, to retain 90% of genetic diversity (heterozygosity) for 100 years (Frankham et al. 2002): this target was devised as a compromise to allow more species to be captive bred while accepting some genetic deterioration in the captive populations. It is a compromise on a compromise as it was originally devised as retaining 90% of genetic diversity for 200 years (Soulé et al. 1986). This is not an appropriate target for species in the wild, especially in the context of their need to adapt to global climate change.
The targets of retaining 90, 95 and 97% of genetic diversity in the CBD discussion material mostly did not specify the duration in years or generations being considered, but from the contexts they appeared to be 2022–2030 (8 years) or 2022–2050 (28 years). However, the impacts of threats to species are more typically evaluated over longer periods, such as the 100 years used in the IUCN Red List Categorization system criterion E for Vulnerable (IUCN 2012) and genetic diversity targets for captive populations of threatened species (Frankham et al. 2010).
I undertook quantitative evaluations of these six scenarios in terms of the loss of genetic diversity over 100 years based on these rates of loss being continued for this duration (Table 1). Other genetic impacts associated with this were also evaluated as described below. The evaluations assume that we are dealing with random mating diploid species, as are most such evaluations.
The means for deriving these extrapolations for genetic diversity are shown for scenario 1, the retention of 90% of genetic diversity for 8 years, as follows:
As 100 years represent 12.5 time-frames of 8 years, the proportion of genetic diversity retained for 100 years (GD100) is:
and
All of the scenarios represent worrying losses of genetic diversity over 100 years (Table 1). However, we need to predict the consequences of these losses on total reproductive fitness and ability to evolve in response to environmental change to appreciate the true impacts on species of following such scenarios.
Consequence of loss of genetic diversity scenarios on inbreeding
In random mating populations, the loss of GD in outbreeding species approximates the inbreeding coefficient (Wright 1969). Most scenarios have reached worrying high levels of inbreeding by year 100 (Table 1). The accumulated mean inbreeding at year 100, is substantially greater than that for the progeny of a full-sib mating in scenarios 3 and 4, near the inbreeding in the progeny of selfing in scenario 2, and near that for the progeny of 2 generations of selfing in scenario 1.
However, it is the consequences on total fitness of these levels of inbreeding that most concern us.
Consequences of loss of genetic diversity scenarios on total fitness (inbreeding depression)
The magnitudes of inbreeding depression (ID) were predicted using the method of Ralls et al. (1988), namely:
where F is the inbreeding coefficient and B the number of haploid lethal equivalents (Morton et al. 1956). This method has been widely used elsewhere, as for example in Frankham et al. (2014), Frankham et al. (2017)). The F values used come from column 5. The B value for total fitness (lifetime reproductive output) of 7.5 lethal equivalents for vertebrates is the median based on the available estimates for vertebrates, while the corresponding B values for outbreeding plants is 3.50 (Frankham et al. 2017, p. 54 and p. 61).
While there are biases in these estimates of inbreeding depression, some are downward and some upwards, so they approximately cancel out. These estimates are biased upwards when inbreeding increases slowly across generations in random mating populations (as here) as natural selection has the opportunity to purge harmful recessive alleles (Day et al. 2003; Reed et al. 2003). However, purging has little effect in small random mating populations representative of many threatened animal populations (Glémin 2003). Conversely, the B values are underestimates as about half of them do not include the effects on offspring fitness of having an inbred versus non-inbred mothers (Frankham et al. 2017).
For those who recall hearing of only 3.14 diploid lethal equivalents for juvenile survival in captive vertebrates (Ralls et al. 1988), the cumulative impacts of inbreeding across reproduction and survival for the whole life cycle are vastly greater than for any single fitness component, and are typically greater in wild than captive habitats (Crnokrak and Roff 1999; O’Grady et al. 2006; Frankham 2015; Frankham et al. 2017).
There are expected to be devastating reductions in total fitness due to inbreeding depression for all six scenarios in vertebrates and plants (Table 1 columns 6 and 7) that will reduce population sizes and increase extinction risks. Even for the least harmful scenario 6, the fitness reductions are very large: the 10% inbreeding coefficient is expected to result in a 54% loss of total fitness in naturally outbreeding vertebrate populations and 30% loss in outbreeding plants.
In addition to inbreeding depression, these small populations are expected to have reduced ability to evolve in response to environmental change, a crucial issue in the context of global climate change (Frankham et al. 2017).
Consequences of loss of genetic diversity scenarios on ability to evolve
In the short term, the major effects of the above scenarios on evolutionary potential are due to reduction in heterozygosity, plus inbreeding depression reducing offspring numbers per female and progeny survival to breeding age, leading to a decline in the selection differential (Frankham et al. 2017, pp. 73–80). The reductions in genetic variation for neutral genetic markers and for fitness should be similar (Kardos et al. 2021, Fig. 1). Consequently, the above scenarios will result in proportionate losses of ability to evolve that are greater than the proportionate losses of genetic diversity (Frankham et al. 2017).
But can we just wait for genetic diversity to be regenerated by mutation?
Recovery of genetic diversity from mutation is far too slow to be an option
Mutation rates are very low so times for mutation to restore genetic diversity are very long. Lande and Barrowclough (1987) estimated them to be hundreds of thousands to millions of generations for single locus genetic diversity and 100 to 1000 of generations for quantitative genetic variation. Empirical evidence accords with the latter prediction (Amador et al. 2010). Consequently, waiting for mutation to restore genetic diversity is not an option in conservation contexts.
What other options do we have for goals for genetic factors in the CBD? Two credible options come to mind:
-
Retaining evolutionary potential in perpetuity
-
Improving population genetic connectivity among fragmented populations
Goal 1: retaining evolutionary potential in perpetuity
Franklin (1980) proposed that an effective size (Ne) of 500 was required to preserve evolutionary potential in perpetuity, based on the equilibrium between neutral mutation and genetic drift for quantitative characters peripheral to fitness. Lande and Barrowclough (1987) reached a similar conclusion, based on a model of mutation, drift, and stabilizing selection.
Consequently, Hoban et al. (2020) and Laikre et al. (2020) proposed “the number of populations (or breeds) with an effective size > 500 compared to the number < 500” as a genetic indicator for CBD. They also specified a proxy for Ne in the absence of genetic data as an adult census size (Nc) of 5,000, based on an average Ne/Nc = 0.1 (Frankham 1995; Palstra and Ruzzante 2008; see also Frankham 2021).
Frankham et al. (2014) re-evaluated the Ne target, based on the accumulated evidence since 1980. They concluded that the objective should be to maintain genetic variation for total reproductive fitness in perpetuity and that the required Ne is at least 1000, based on empirical and theoretical work.
This is a credible indicator, but using effective population size in a CBD indicator is problematical as:
-
Ne is far too complex for non-geneticists, as the literature is extraordinarily complex and confusing, such that even specialist evolutionary geneticists make mistakes (e.g. Frankham 1995; Hoban et al. 2020; see Frankham 2021). For example, there are many different variables called Ne and they differ in magnitude (Frankham 1995; Wang et al. 2016; Ryman et al. 2019).
-
In practice, this indicator will revert to Nc in the vast majority of cases as appropriate estimates of Ne (multigenerational ones) are available for few species (Frankham 2021), while Nc estimates are available for many species (e.g. IUCN 2022). Further, very few appropriate genetic estimates of Ne are now being undertaken due to the need for samples separated by several generations.
These issues can be largely overcome by using the median estimate of Ne/Nc of ~ 0.1 (Frankham 1995, 2021) to convert the indicator from an Ne of 1,000 to a census size of 10,000, following the approach of Hoban et al. (2020) and Laikre et al. (2020). However, Laikre et al (2021) argued for the retention of Ne with a default Nc of ten times this value when Ne is not available. In part, this was based on the existence of variation in Ne/Nc ratios among species. By contrast, Hoban et al. (2021) concluded that “In the absence of such information (on species specific Ne), the rule of thumb of Ne/Nc = 0.1 is an empirically justified, conservative threshold for many if not most organisms.” I remain unconvinced of the desirability of specifying Ne, as much of the influential audience for implementation of the CBD consists of non-geneticists, especially bureaucrats and politicians from counties around the world, and for whom Ne will represent unwarranted and unwelcome complexity. I suggest we follow the lead of economists who use simple, easily measured indicators such as gross domestic product, unemployment rate, etc., and who have achieved substantial political influence.
Goal 2: improving population genetic connectivity
Most species have fragmented distributions, many with small isolated populations that have low genetic diversity, are inbred, and have reduced fitness and ability to evolve (Frankham et al. 2017; Frankham et al. 2019). An estimated ~ 1.4 million isolated populations of threatened species are suffering genetic erosion, and for non-threatened plus threatened species the number climbs to ~ 150 million isolated populations with genetic problems (Frankham et al. 2017).
Genetic management of fragmented populations has been described as one of the most important, largely unaddressed issues in all of conservation biology (Frankham 2010a, 2010b). A major component of this management is to increase gene flow in cases where it has ceased or become inadequate so that genetic problems associated with small isolated populations are prevented or reversed (Frankham et al. 2017). Gene flow can be increased by:
-
Increasing population sizes where populations are sufficiently close for this to increase gene flow
-
Removing barriers to gene flow
-
Building habit corridors
-
Building wildlife underpasses and overpasses
-
Moving individuals or gametes between populations (genetic rescue attempts)
Increasing population size
Increasing population size is expected to increase gene flow if the rate of migration is unchanged. However, land clearing often creates habitat fragments in an inhospitable matrix, resulting in reduced rates of gene flow (Frankham et al. 2017). Further, habitat clearing typically reduces the carrying capacity for populations, such that they may not be able to support increased population size unless habitat restoration is undertaken.
Removing barriers to gene flow
Humans have in many cases inserted barriers to gene flow, such as dams/weirs, fences, and roads. Removing these can re-establish gene flow (Frankham et al. 2017). For example, many dams and weirs have been removed from rivers, presumably restoring gene flow.
Building habitat corridors
Adding strips of suitable habitat between isolated populations is another means to restore gene flow (Frankham et al. 2017). This depends on the corridors being used by the target species.
Building wildlife underpasses and overpasses
In a similar manner building wildlife overpasses or underpasses can improve gene flow where it is inadequate (Frankham et al. 2017).
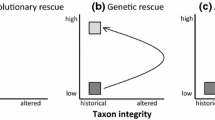
Genetic rescue attempts
If the four actions above are not feasible, as may frequently be the case, the remaining option is human assisted movement of individuals or gametes to re-establish gene flow. In the past, genetic rescue attempts were impeded by concerns that gene flow would lead to harmful effects (outbreeding depression) (Edmands 2007). However, the causes of outbreeding depression are known and means to predict its risks have been devised and validated (Frankham et al. 2011; Frankham 2015).
Genetic rescue attempts have resulted in large and consistently beneficial effects that persist over generations in outbreeding species (Frankham 2015, 2016; Frankham et al. 2019). Outcrossing of inbred populations resulted in beneficial effects in 92.9% of 156 cases screened as having a low risk of outbreeding depression. The median increase in composite fitness (combined fecundity and survival) following outcrossing was 148% in wild/stressful environments and 45% in captive/benign ones. Consequently, genetic rescues are a highly effective genetic management tool.
Proposed indicators of improved population genetic connectivity for the CBD
People discussing non-genetic aspects of the CBD often specify options to alleviate their threats and reverse declines (Maron et al. 2021). This is not currently a part of the proposal to strengthen the indicators for genetic connectivity in the CBD but is equally needed there. It also addresses the issue of offering hope and good news stories.
Each of the five items above are potential CBD indicators of improvements in genetic connectivity. However, for each to be credible it needs to be established by genetic monitoring that the genetic connectivity was initially inadequate and that the action resulted in improved genetic connectivity (Allendorf et al. 2022).
I recommend that the cumulative total of cases of credible improvements in genetic connectivity from these five items be included as a genetic connectivity indicator in the CBD. They are easily understood and measured. Baseline lists of number of prior global genetic rescue attempts already exist in Frankham et al. (2011, Table S1), and Frankham et al. (2017, Table 1.1), and a further update is projected to appear in a forthcoming textbook. The cumulative global numbers of genetic rescues rose from 19 to 29 between 2011 and 2017 and were approximately 34 by 2021 (Frankham et al. unpublished). I am confident that there will be an increase in the number of these indicators that will lead to good news stories in 2030 and 2050.
Conclusion
All six percentage-based genetic diversity target scenarios result in harmful losses of genetic diversity, increased inbreeding, major losses of fitness, and reduced ability to evolve. Consequently, none of these scenarios should be used in the CBD revision. In general, conservation targets that specify loss of genetic diversity are goals for harming species and are inappropriate in conservation contexts.
However, genetic goals, targets and indicators can and should be incorporated in biodiversity monitoring more generally, and in the CBD specifically. Two alternative indicators are proposed, namely the proportion of species and their populations being maintained at sizes sufficient to retain evolutionary potential in perpetuity, and the number of species where population connectivity has been improved.
In addition, the wording of CBD Goals, Milestones, and Targets must be specific enough regarding the conservation of genetic diversity for these indicators to be relevant for countries to report. The Goals, Milestones, and Targets should mention elements such as maintaining sufficiently large populations, sufficient and appropriate genetic exchange, and active monitoring and management of genetic diversity, as well as no loss of populations.
Data availability
I conceived the ideas, prepared the Table based on my computations, and wrote the paper. All associated data are included in the paper.
References
Allendorf FW et al (2022) Conservation and the genomics of populations, 3rd edn. Oxford University Press, Oxford
Amador C et al (2010) Regeneration of the variance of metric traits by spontaneous mutation in a Drosophila population. Genet Res 92:91–102
Crnokrak P, Roff DA (1999) Inbreeding depression in the wild. Heredity 83:260–270
Day SB, Bryant EH, Meffert LM (2003) The influence of variable rates of inbreeding on fitness, environmental responsiveness, and evolutionary potential. Evolution 57:1314–1324
Edmands S (2007) Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding depression for conservation and management. Mol Ecol 16:463–475
Frankham R (1995) Effective population size/adult population size ratios in wildlife: a review. Genet Res 66:95–107
Frankham R (2010a) Where are we in conservation genetics and where do we need to go? Conserv Genet 11:661–663
Frankham R (2010b) Challenges and opportunities of genetic applications in biological conservation. Biol Conserv 143:1919–1927
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol 24:2610–2618
Frankham R (2016) Genetic rescue benefits persist to at least the F3 generation, based on a meta-analysis. Biol Conserv 195:33–36
Frankham R (2021) Suggested improvements to proposed genetic indicator for CBD. Conserv Genet 22:531–532
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Frankham R, Ballou JD, Briscoe DA (2010) Introduction to conservation genetics, 2nd edn. Cambridge University Press, Cambridge
Frankham R et al (2011) Predicting the probability of outbreeding depression. Conserv Biol 25:465–475
Frankham R, Bradshaw CJA, Brook BW (2014) Genetics in conservation management: revised recommendations for the 50/500 rules, Red List criteria and population viability analyses. Biol Conserv 170:56–63
Frankham R et al (2017) Genetic management of fragmented animal and plant populations. Oxford University Press, Oxford
Frankham R et al (2019) A practical guide for genetic management of fragmented animal and plant populations. Oxford University Press, Oxford
Franklin IR (1980) Evolutionary change in small populations. In: Soule ME, Wilcox BA (eds) Conservation biology: an evolutionary-ecological perspective. Sinauer, Sunderland, pp 135–150
Glémin S (2003) How are deleterious mutations purged? drift versus nonrandom mating. Evolution 57:2678–2687
Hoban S et al (2020) Genetic diversity targets and indicators in the CBD post-2020 global biodiversity framework must be improved. Biol Conserv 248:108564
Hoban S et al (2021) Effective population size remains a suitable, pragmatic indicator of genetic diversity for all species, including forest trees. Biol Conserv 253:108906
IUCN (2012) IUCN Red List categories and criteria version 3.1., 2nd edn. Gland, Cambridge
IUCN (2022) IUCN Red List of threatened species. Available at http://www.redlist.org/. Accessed 26 May 2022
Kardos M et al (2021) The crucial role of genome-wide genetic variation in conservation. Proc Natl Acad Sci USA 118:e2104642118
Laikre L et al (2020) Post-2020 goals overlook genetic diversity. Science 367:1083–1085
Laikre L et al (2021) Authors’ reply to letter to the editor: continued improvement to genetic diversity indicator for CBD. Conserv Genet 22:533–536
Lande R, Barrowclough GF (1987) Effective population size, genetic variation, and their use in population management. In: Soulé ME (ed) Viable populations for conservation. Cambridge University Press, Cambridge, pp 87–123
Maron M et al (2021) Setting robust biodiversity goals. Conserv Lett 14:e12816
Morton NE, Crow JF, Muller HJ (1956) An estimate of the mutational damage in man from data on consanguineous marriages. Proc Natl Acad Sci USA 42:855–863
O’Grady JJ et al (2006) Realistic levels of inbreeding depression strongly affect extinction risk in wild populations. Biol Conserv 133:42–51
Palstra FP, Ruzzante DE (2008) Genetic estimates of contemporary effective population size: what can they tell us about the importance of genetic stochasticity for wild population persistence? Mol Ecol 17:3428–3447
Ralls K, Ballou JD, Templeton A (1988) Estimates of lethal equivalents and the cost of inbreeding in mammals. Conserv Biol 2:185–193
Reed DH et al (2003) Inbreeding and extinction: effects of rate of inbreeding. Conserv Genet 4:405–410
Ryman N, Laikre L, Hössjer O (2019) Do estimates of contemporary effective population size tell us what we want to know? Mol Ecol 28:1904–1918
Soulé ME et al (1986) The millenium ark: how long a voyage, how many staterooms, how many passengers? Zoo Biol 5:101–113
Wang J, Santiago E, Caballero A (2016) Prediction and estimation of effective population size. Heredity 117:193–206
Wright S (1969) Evolution and the genetics of populations 2 the theory of gene frequencies. University of Chicago Press, Chicago
Acknowledgements
I thank Craig Moritz for inviting me to contribute to a meeting on genetic revisions to the CBD, Caroline Lees and Onnie Byers for inviting me to contribute to the IUCN taskforce on genetic targets and goals for the revised CBD as a representative of CPSG, to other members of that task force for feedback on the position paper, and to Sean Hoban for suggesting that I convert it into a Perspective for Conservation Genetics. I am grateful to Sean Hoban, Joachim Mergeay, and two anonymous reviewers for their helpful comments.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. The author declares that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
I conceived the ideas, prepared the Table based on my computations, and wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The author has no relevant financial or non-financial interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Frankham, R. Evaluation of proposed genetic goals and targets for the Convention on Biological Diversity. Conserv Genet 23, 865–870 (2022). https://doi.org/10.1007/s10592-022-01459-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-022-01459-1