Abstract
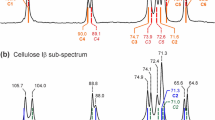
Solid-state 13C nuclear magnetic resonance (NMR) spectroscopy is a powerful tool for identifying the various allomorphs of cellulose, for example quantifying the proportions of cellulose Iα and Iβ allomorphs in the crystalline domains of various native celluloses. While marine invertebrate animals known as tunicates produce nearly pure cellulose Iβ, solid-state NMR reveals that bacterial cellulose is dominated by the Iα allomorph, but also has additional signals arising from a secondary crystalline form that are normally attributed to cellulose Iβ. However, in this paper we show that the 13C chemical shifts and correlation patterns in 2D NMR spectra for this secondary crystalline form in bacterial cellulose are not the same as those found in cellulose Iβ spectra reported for tunicate cellulose. In order to reach this conclusion, it was necessary to ensure all spectra were correctly set to the same chemical shift scale. In doing so, it became apparent there were spectral features for bacterial cellulose that were entirely consistent with previously reported spectra of bacterial cellulose but did not match those of the Iβ allomorph found in tunicate cellulose. Through a careful and detailed analysis of the 1D and 2D NMR spectra of three 13C-enriched bacterial cellulose samples, two sets of correlated 13C chemical shifts for this secondary crystalline form were identified. The fact that these chemical shifts and correlation patterns are different than those of tunicate cellulose suggests that there exists some sort of structural variation from the Iβ allomorph in bacterial cellulose since the 13C NMR spectrum closely resembles, but is not identical to, the 13C spectrum of the Iβ allomorph found in tunicate cellulose.






Similar content being viewed by others
Data availability
The Supporting Information provides full details on the models used to calculate and fit the experimental 13C NMR spectra of the bacterial cellulose samples. It also provides comparisons of the 13C chemical shifts for the tunicate cellulose Iβ allomorph to the experimental 1D and 2D NMR spectra. The Mathematica notebook used for fitting the 1D and 2D spectra, as well as the raw data for each spectrum, is also provided in the Supporting Information.
References
Arashida T, Ishino T, Kai A et al (1993) Biosynthesis of cellulose from culture media containing 13C-labeled glucose as a carbon source. J Carbohydr Chem 12:641–649. https://doi.org/10.1080/07328309308019413
Atalla RH, Vanderhart DL (1984) Native cellulose: a composite of two distinct crystalline forms. Science 223:283–285. https://doi.org/10.1126/science.223.4633.283
Atalla RH, Vanderhart DL (1999) The role of solid state 13C NMR spectroscopy in studies of the nature of native celluloses. Solid State Nucl Magn Reson 15:1–19. https://doi.org/10.1016/S0926-2040(99)00042-9
Belton PS, Tanner SF, Cartier N, Chanzy H (1989) High-resolution solid-state 13C nuclear magnetic resonance spectroscopy of tunicin, an animal cellulose. Macromolecules 22:1615–1617. https://doi.org/10.1021/ma00194a019
Brouwer DH, Mikolajewski JG (2023) Resolving the discrepancies in reported 13C solid state NMR chemical shifts for native celluloses. Cellulose 30:4827–4839. https://doi.org/10.1007/s10570-023-05186-9
Carravetta M, Edén M, Zhao X et al (2000) Symmetry principles for the design of radiofrequency pulse sequences in the nuclear magnetic resonance of rotating solids. Chem Phys Lett 321:205–215. https://doi.org/10.1016/S0009-2614(00)00340-7
Fink H-P, Purz HJ, Bohn A, Kunze J (1997) Investigation of the supramolecular structure of never dried bacterial cellulose. In: Macromolecular symposia vol 120, pp 207–217. https://doi.org/10.1002/masy.19971200121
French AD (2014) Idealized powder diffraction patterns for cellulose allomorphs. Cellulose 21:885–896. https://doi.org/10.1007/s10570-013-0030-4
Fung BM, Khitrin AK, Ermolaev K (2000) An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson 142:97–101. https://doi.org/10.1006/jmre.1999.1896
Hestrin S, Schramm M (1954) Synthesis of cellulose by Acetobacter xylinum. II. Preparation of freeze-dried cells capable of polymerizing glucose to cellulose. Biochem J 58:345–352. https://doi.org/10.1042/bj0580345
Horii F, Hirai A, Kitamaru R (1984) CP/MAS carbon-13 NMR study of spin relaxation phenomena of cellulose containing crystalline and noncrystalline components. J Carbohydr Chem 3:641–662. https://doi.org/10.1080/07328308408057922
Horii F, Hirai A, Kitamaru R (1987) CP/MAS 13C NMR spectra of the crystalline components of native celluloses. Macromolecules 20:2117–2120. https://doi.org/10.1021/ma00175a012
Idström A, Schantz S, Sundberg J et al (2016) 13C NMR assignments of regenerated cellulose from solid-state 2D NMR spectroscopy. Carbohydr Polym 151:480–487. https://doi.org/10.1016/j.carbpol.2016.05.107
Kai A, Arashida T, Hatanaka K et al (1994) Analysis of the biosynthetic process of cellulose and curdlan using 13C-labeled glucoses. Carbohydr Polym 23:235–239. https://doi.org/10.1016/0144-8617(94)90184-8
Kang X, Kirui A, Dickwella Widanage MC et al (2019) Lignin-polysaccharide interactions in plant secondary cell walls revealed by solid-state NMR. Nat Commun 10:1–9. https://doi.org/10.1038/s41467-018-08252-0
Kirui A, Zhao W, Deligey F et al (2022) Carbohydrate-aromatic interface and molecular architecture of lignocellulose. Nat Commun 13:1–12. https://doi.org/10.1038/s41467-022-28165-3
Koch F-T, Prieß W, Witter R, Sternberg U (2000) Calculation of solid-state 13C NMR spectra of cellulose Iα, Iβ and II using a semi-empirical approach and molecular dynamics. Macromol Chem Phys 201:1930–1939. https://doi.org/10.1002/1521-3935(20001001)201:15%3c1930::AID-MACP1930%3e3.0.CO;2-7
Kono H, Yunoki S, Shikano T et al (2002) CP/MAS C-13 NMR study of cellulose and cellulose derivatives. 1. Complete assignment of the CP/MAS C-13 NMR spectrum of the native cellulose. J Am Chem Soc 124:7506–7511. https://doi.org/10.1021/Ja010704o
Kono H, Erata T, Takai M (2003a) Determination of the through-bond carbon-carbon and carbon-proton connectivities of the native celluloses in the solid state. Macromolecules 36:5131–5138. https://doi.org/10.1021/ma021769u
Kono H, Erata T, Takai M (2003b) Complete assignment of the CP/MAS 13C NMR spectrum of cellulose IIII. Macromolecules 36:3589–3592. https://doi.org/10.1021/ma021015f
Kono H, Numata Y, Erata T, Takai M (2004) 13C and 1H resonance assignment of mercerized cellulose II by two-dimensional MAS NMR spectroscopies. Macromolecules 37:5310–5316. https://doi.org/10.1021/ma030465k
Langan P, Nishiyama Y, Chanzy H (2001) X-ray structure of mercerized cellulose II at 1 Å resolution. Biomacromolecules 2:410–416. https://doi.org/10.1021/bm005612q
Larsson PT, Westermark U, Iversen T (1995) Determination of the cellulose Iα allomorph content in a tunicate cellulose by CP/MAS 13C-NMR spectroscopy. Carbohydr Res 278:339–343. https://doi.org/10.1016/0008-6215(95)00248-0
Malm E, Bulone V, Wickholm K et al (2010) The surface structure of well-ordered native cellulose fibrils in contact with water. Carbohydr Res 345:97–100. https://doi.org/10.1016/j.carres.2009.10.020
Morcombe CR, Zilm KW (2003) Chemical shift referencing in MAS solid state NMR. J Magn Reson 162:479–486. https://doi.org/10.1016/S1090-7807(03)00082-X
Mori T, Chikayama E, Tsuboi Y et al (2012) Exploring the conformational space of amorphous cellulose using NMR chemical shifts. Carbohydr Polym 90:1197–1203. https://doi.org/10.1016/j.carbpol.2012.06.027
Nishiyama Y, Langan P, Chanzy H (2002) Crystal structure and hydrogen-bonding system in cellulose Iβ from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc 124:9074–9082. https://doi.org/10.1021/ja0257319
Nishiyama Y, Sugiyama J, Chanzy H, Langan P (2003) Crystal structure and hydrogen bonding system in cellulose Iα from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc 125:14300–14306. https://doi.org/10.1021/ja037055w
O’Sullivan AC (1997) Cellulose: the structure slowly unravels. Cellulose 4:173–207. https://doi.org/10.1023/A:1018431705579
Phyo P, Wang T, Yang Y et al (2018) Direct determination of hydroxymethyl conformations of plant cell wall cellulose using 1H polarization transfer solid-state NMR. Biomacromolecules 19:1485–1497. https://doi.org/10.1021/acs.biomac.8b00039
Ross P, Mayer R, Benziman M (1991) Cellulose biosynthesis and function in bacteria. Microbiol Rev 55:35–58. https://doi.org/10.1128/mr.55.1.35-58.1991
States DJ, Haberkorn RA, Ruben DJ (1982) A two-dimensional nuclear overhauser experiment with pure absorption phase in four quadrants. J Magn Reson 48:286–292. https://doi.org/10.1016/0022-2364(82)90279-7
Sternberg U, Koch F-T, Priess W, Witter R (2003) Crystal structure refinements of cellulose allomorphs using solid state 13C chemical shifts. Cellulose 10:189–199. https://doi.org/10.1023/A:1025185416154
VanderHart D, Atalla R (1987) Further carbon-13 NMR evidence for the coexistence of two crystalline forms in native celluloses. ACS Symp Ser 340:88–118. https://doi.org/10.1021/bk-1987-0340.ch005
VanderHart DL, Atalla RH (1984) Studies of microstructure in native celluloses using solid-state 13C NMR. Macromolecules 17:1465–1472. https://doi.org/10.1021/ma00138a009
Wada M, Chanzy H, Nishiyama Y, Langan P (2004a) Cellulose IIII crystal structure and hydrogen bonding by synchrotron X-ray and neutron fiber diffraction. Macromolecules 37:8548–8555. https://doi.org/10.1021/ma0485585
Wada M, Heux L, Sugiyama J (2004b) Polymorphism of cellulose I family: Reinvestigation of cellulose IVl. Biomacromol 5:1385–1391. https://doi.org/10.1021/bm0345357
Wada M, Heux L, Nishiyama Y, Langan P (2009) X-ray crystallographic, scanning microprobe X-ray diffraction, and cross-polarized/magic angle spinning 13C NMR studies of the structure of cellulose IIIII. Biomacromol 10:302–309. https://doi.org/10.1021/bm8010227
Wang T, Yang H, Kubicki JD, Hong M (2016) Cellulose structural polymorphism in plant primary cell walls investigated by high-field 2D solid-state NMR spectroscopy and density functional theory calculations. Biomacromolecules 17:2210–2222. https://doi.org/10.1021/acs.biomac.6b00441
Wolfram Research Inc (2021) Mathematica. version 13.0
Yates JR, Pham TN, Pickard CJ et al (2005) An investigation of weak CH⋯O hydrogen bonds in maltose anomers by a combination of calculation and experimental solid-state NMR spectroscopy. J Am Chem Soc 127:10216–10220. https://doi.org/10.1021/ja051019a
Acknowledgments
The authors thank the Department of Chemistry and Chemical Biology at McMaster University for access to the 850 MHz NMR spectrometer.
Funding
This work was supported by the Natural Sciences and Engineering Research Council (NSERC) of Canada in the form of a Discovery Grant (funding reference number 03836) and an Undergraduate Student Research Award.
Author information
Authors and Affiliations
Contributions
Material preparation was carried out by JM. Data collection, analysis, and writing of the manuscript were carried out by DB. Both authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Brouwer, D.H., Mikolajewski, J.G. Solid-state NMR reveals a structural variation from the Iβ polymorphic form in bacterial cellulose. Cellulose 30, 11341–11356 (2023). https://doi.org/10.1007/s10570-023-05577-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10570-023-05577-y