Abstract
Surgical resection, when combined with chemotherapy, has been shown to significantly improve the survival rate of patients with pancreatic ductal adenocarcinoma (PDAC). However, this treatment option is only feasible for a fraction of patients, as more than 50% of cases are diagnosed with metastasis. The multifaceted process of metastasis is still not fully understood, but recent data suggest that transcriptional and epigenetic plasticity play significant roles. Interfering with epigenetic reprogramming can potentially control the adaptive processes responsible for metastatic progression and therapy resistance, thereby enhancing treatment responses and preventing recurrence. This review will focus on the relevance of histone-modifying enzymes in pancreatic cancer, specifically on their impact on the metastatic cascade. Additionally, it will also provide a brief update on the current clinical developments in epigenetic therapies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Background
Pancreatic ductal adenocarcinoma (PDAC) is the third leading cause of cancer-related death in the USA and has the lowest 5-year survival rate of all cancer types, with only 12% [1]. The current standard of care for most patients involves modestly effective combination chemotherapies, such as a modified FOLFIRINOX regimen or gemcitabine and nanoparticle albumin-bound paclitaxel [2]. By the time of diagnosis, around 50% of patients have already developed metastasis, while around 30 to 35% have locally advanced, unresectable tumors. Only 10–20% of patients harbor localized tumors, which allows curative-intent resections in the context of neo-adjuvant/adjuvant therapies [1,2,3]. However, it is important to note that even resectable PDACs are likely to be disseminated and should be treated as a systemic disease. Genetically engineered mouse models have shown inflammation-driven epithelial to mesenchymal transition (EMT) and dissemination early in the disease [4]. Furthermore, the metastatic probability of a 2-cm-sized PDAC was calculated to be 73%, and 94% for 3-cm-sized cancer, respectively. Considering the average diameter of PDAC at surgery is 3–4 cm underscores the need for neo-adjuvant/adjuvant therapies even for resectable patients [5]. Furthermore, these data highlight the demand for a detailed understanding of the metastatic cascade.
Tumor initiation in PDAC is predominantly caused by KRAS mutations, occurring in approximately 90% of patients. In addition to KRAS, major genetic alterations occur in TP53, CDKN2A, and SMAD4 in up to 80%, 26%, and 25% of patients, respectively [6,7,8]. The sequencing of primary tumors and matched metastatic tissues has demonstrated high concordance of driver gene mutations [9, 10]. However, increased gene dose of the KRAS oncogene [11, 12] as well as the amplification or activation of MYC [13, 14] have been linked to tumor progression and metastasis. When considering frequencies, e.g., major KRAS imbalances in favor of the mutant allele occur in 4% of primary tumors compared to 29% in metastatic disease [12], it is probable that multiple pathways contribute. Furthermore, the described concordance of mutations in metastasizing and non-metastasizing PDACs underscores the contribution of non-genetic, transcriptional reprogramming and epigenetic plasticity to the metastatic cascade of PDAC. Early in carcinogenesis, differential chromatin modules identify heterogeneous cell-states primed for later emerging tumor cell fates [15]. This emphasizes the significance of epigenetic plasticity in the biology of PDAC. Consistently, large chromatin domains are reprogrammed during tumor progression [10], and a growing set of transcription factors (TF), such as Forkhead family TFs [16, 17], BLIMP1 [18], RUNX3 [19], ZEB1 [20], PRRX1 [21], FOSL1 [22, 23], or deltaNp63 [24, 25], have been associated with dedifferentiation and metastasis.
The liver and lungs are the most commonly affected distant organ sites where metastasis of PDAC occurs. The molecular patterns responsible for this organ site specificity are currently under investigation [7, 26,27,28]. Recent efforts to subtype PDAC have identified two classical (A/B) and two basal-like (A/B) subtypes, as well as a hybrid subtype [12]. The classical subtypes are characterized by a slightly better prognosis. These subtypes exhibit an enriched expression of the pancreatic differentiation factor GATA6 and an increased occurrence of SMAD4 alterations. In contrast, basal-like subtypes are characterized by an enrichment of EMT and TGF-beta signatures as well as an increased occurrence of TP53 mutations and complete loss of CDKN2A [12]. The most aggressive and chemoresistant subtype, basal-like A, displays the highest frequency of intact SMAD4 [12]. As basal-like A PDACs exhibit a higher probability of metastasis and are enriched in EMT and TGF-beta signatures, it raises the question of how the tumor-suppressive arm of TGF-beta signaling is overcome in SMAD4 wild-type PDACs, a question addressed by several recent studies [29,30,31]. While EMT and TGF-beta signaling are tightly linked and promote a more aggressive phenotype, an increased understanding of cellular plasticity has promoted the concept of partial EMT (p-EMT) as a more plastic, reversible morphological state [32]. The characteristics of p-EMT fundamentally differ from the classical EMT processes. Particularly, p-EMT might be mediated independently of TGF-beta signaling. Observations of circulating tumor cells (CTC) also discovered that cells in a p-EMT state often disseminate in clusters whereas fully mesenchymal cells mostly travel as individual cells in the vascular system. On closer inspection, pancreatic cancer patients with a higher number of CTCs in the p-EMT state displayed a shorter progression-free survival [33].
The cellular, transcriptional, and epigenetic plasticity of cancer cells [34, 35] is especially relevant in PDAC [36]. Cellular plasticity processes induced by intracellular or extracellular cues can occur within just a few days in vivo, completely changing the metastatic capabilities. In murine autochthonous PDAC, genetic manipulation of the bone morphogenetic protein (BMP) signaling inhibitor Gremlin 1 (GREM1) in vivo converted epithelial differentiated PDACs to undifferentiated mesenchymal PDAC, which was linked to a distinct increase in metastatic frequency [37]. Mesenchymal cells produce GREM1, which acts in a paracrine fashion to block BMP signaling and the induction of the EMT transcription factors Snail family transcriptional repressor 1/2 (SNAI1/2) in epithelial cells. These findings illustrate how PDACs use signaling-linked transcriptional reprogramming to shape intra-tumoral heterogeneity and metastasis [37]. Additionally, these findings suggest that tumor cells require highly adaptable and fluidic capabilities to pass through the complete metastatic cascade (Fig. 1), which is known to be tightly controlled by transcriptional networks and the epigenetic machinery [10, 38,39,40,41]. The support for this note is based on an additional study that aimed to identify metastasis drivers in a large cohort of molecularly characterized cancer patients. The study involved 1779 PDAC patients and compared primary tumors with metastatic tumors. Alongside CDKN2A and SMAD4 deletions, the TGF-beta signaling pathway, cell-cycle signatures, and the epigenetic pathway were linked to metastasis in PDAC [7]. These epigenetic pathways encompass a variety of mechanisms that contribute to cellular plasticity and metastasis. For instance, elegant studies identified hypomethylated DNA signatures in PDAC cells that coordinate the reorganization of the microenvironment via Interferon-alpha (IFNα) signaling and link DNA methylation to the more aggressive basal-like subtype [42]. Beyond DNA methylation, the genome’s three-dimensional architecture, non-coding RNAs, and histone modifications present other relevant ways of epigenetic contribution to plasticity [43]. Here we focus on recent insights into the contribution of histone-modifying enzymes to metastasis and their potential as a therapeutic target. We will spotlight lysine modifiers.
PDAC cells undergo chromatin remodeling during metastatic processes. Disseminating tumor cells constantly change their chromatin landscape to adapt to the challenges during the metastatic process. These include EMT, migration and invasion, intravasation, anchorage-independent survival in the circulation, extravasation, nesting and dormancy, MET, proliferation, and distant organ colonization
2 Histone modifiers in PDAC metastasis
The dissemination of tumor cells is a complex process that involves several stages in the formation of metastases and requires continuous adaptation [35] (Fig. 1). Starting with challenges in nutrient supply during tumor growth, disseminating cells are forced to survive the lack of adhesion during their travel to distant organs while constantly evading immune surveillance. Once tumor cells arrive at a distant organ site, they must re-establish cell growth in a completely new (micro-) environment [3, 41]. Unlike fixed genomic alterations, the reversibility of histone marks grants cancer cells the capability to undergo dynamic shifts in the epigenetic landscape, which is relevant to cope with challenges posed by the metastatic cascade [41]. Histone-modifying enzymes, such as histone deacetylases (HDACs), histone acetyltransferases (HATs), as well as lysine methyltransferases (KMTs) and lysine demethylases (KDMs), play crucial roles in conferring adaptability to disseminating cells. Notably, the reprogramming of enhancers by epigenetic modifications at histones during metastatic transition and as means of drug resistance is a commonly observed phenomenon [16, 44]. This is furthermore illustrated by a previous study, which demonstrated the linkage between metabolic changes in primary PDAC tumors and metastatic sites, mediated by a global reorganization of the epigenetic landscape [10]. Additionally, the study showed the collective changes in histone methylation and acetylation as well as their interdependence, demonstrating the relevance of the epigenetic machinery for PDAC metastasis.
Histone modifications, such as acetylation or methylation, can coordinate diverse transcriptional dynamics depending on their genomic location and the modified histone residue. For example, H3K4 trimethylation (H3K4me3) along with H3K27 acetylation (H3K27ac) are typically observed at open and active promoter sites [45,46,47,48,49,50]. On the other hand, H3K27 trimethylation (H3K27me3) signifies heterochromatin and causes transcriptional repression (Fig. 2a) [51]. H3K27ac at promotor and enhancer sites can lead to the binding of bromodomain and extraterminal domain (BET) proteins that act as scaffolds for the recruitment of the transcription elongation machinery as well as transcription factors [52,53,54]. The family of BET proteins, often referred to as “epigenetic readers,” includes bromodomain-containing proteins (BRD) 2-4 as well as the testis-specific BRDT. Particularly, BRD4 has been well characterized as it recruits positive transcriptional elongation factor (P-TEFb) to acetylated transcriptional start sites and promotes transcriptional elongation [55]. Numerous studies showed a causal relationship with expression of oncogene c-MYC and BRD4, including pancreatic cancer [56]. For instance, inhibition of BRD4 by shRNA or BET-inhibitor (BETi) JQ1 reduced MYC protein levels in PDAC cell lines [56]. Additionally, JQ1 effectively inhibited tumor growth in primary pancreatic cancer xenografts in MYC high groups, providing a stratification strategy for MYC-dependent tumors [57]. Furthermore, BRD4 is frequently recruited by enhancer-binding factors such as yes-associated protein (YAP)/PDZ-binding motif (TAZ) to enable enhancer-promoter interaction in malignancies [58]. Recent studies identified the transcriptional control of receptor tyrosine kinase-like orphan receptor 1 (ROR1) via YAP-BRD4 enhancer interactions and associated high ROR1 expression with a partial-EMT state. In this study, ROR1-induced tumorigenesis and its knockdown reduced metastasis formation in orthotopically transplanted mice [59]. BET protein-dependent enhancer-promoter interaction also influences subtype identities in PDAC. Elegant studies discovered that BET family member bromodomain containing 4 (BRD4) maintained cJUN expression which resulted in the recruitment of TNF-alpha positive macrophages via the BRD4-cJUN/AP1-CCL2 axis and subsequent TNF-alpha-mediated classical-to-basal subtype switch [60]. In addition, cJUN collaborates with progenitor transcription factors, like Sox2, Sox5, Prrx1, Twist2, or Nr2f2 to overcome Yap dependency [61], which is associated with an EMT state that is different from classical TGF-beta-driven EMT. YAP-independent PDAC can be targeted with BETi, and therefore, the combination of BETi with YAPi exhibits synergism [61].
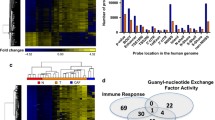
Epigenetic readers and writers in PDAC. A HATs and KMTs induce euchromatin by acetylation of H3K27 and methylation of H3K4. HDACs and KMT deacetylate and methylate H3K27 to induce heterochromatin. KDMs remove methylations at H3K4 or H3K27, respectively. B (Left): 15% (n=266) of PDAC patients in the MSK MetTropsim PDAC cohort (n=1779) display mutations in histone-modifying enzymes (Right): Upset-plot displaying the number of patients with single and multiple (connected dots) mutations in histone-modifying enzymes. Data and plots were in part extracted from cBioPortal [73, 74]
To physically connect promoter- and enhancer-associated proteins, promoter-enhancer interactions often require scaffolding proteins to assemble these factors located at different genomic regions. A frequently mutated scaffold protein, menin (MEN1), has been described as a tumor suppressor, and its inactivating mutations induce endocrine tumors including pancreatic neuroendocrine tumors (PanNETs). In PanNETs, nearly 40% of patients harbor MEN1 mutations and it is the most commonly mutated gene [62, 63]. Additionally, reports show that non-mutated PanNETs often lack MEN1 expression [64, 65]. In this regard, a recent study identified the degradation of MEN1 via the E3 ligase Cullin4b. Inhibition of MEN1 neddylation or knockdown of CUL4 substrate receptor DCAF7 induced MEN1 protein accumulation and resulted in impeded malignant biological behaviors in vitro and reduced tumor volume in vivo [66]. In PDAC, on the other hand, MEN1 is rarely mutated and seems to play a less important role in tumor development. Nevertheless, novel reports show that MEN1 interacts with histone deacetylases to repress CCAAT enhancer binding protein beta (C/EBPB) expression during TGF-beta signaling, inducing EMT and preventing C/EBPB-mediated tumor suppression [30].
Several attempts have been made to associate epigenetic patterns with PDAC subtypes [12, 40]. Lomberk et al. conducted an analysis of various chromatin states and linked these epigenetic clusters to the basal-like and classical PDAC subtype. Their study confirmed the alteration of specific enhancer and promoter regions in these subtypes. As anticipated, functional analysis of the epigenetic control regions demonstrated programs connected to pancreatic development and Ras signaling in the classical subtype. In basal-like PDAC, enhancer controlled programs for epidermal growth factor receptor (EGFR) and phosphatidylinositol 3-kinase (PI3K)-signaling as well as EMT-related pathways such as TGF-beta were detected. In addition, networks involved in cell differentiation and proliferation were linked to the epigenomic cluster matching basal-like PDACs [12, 40].
Apart from the modified epigenetic landscape in PDAC subtypes, a significant proportion of PDAC cases exhibit mutations in histone modifiers. A recent study revealed that the collective frequency of mutations in histone-modifying enzymes in PDAC patients surpasses 14%, emphasizing their importance (Fig. 2b) [7]. Several other studies have indicated a mutational frequency of up to 24% in various histone modifiers, with the most commonly mutated genes being the histone demethylase KDM6A as well as the histone methyl transferases SETD2, KMT2C (MLL3), KMT2D (MLL3) [6, 67, 68]. Other histone modifiers were less frequently affected (Fig. 2b). Beyond mutations, expression patterns of histone modifiers have also been intensively researched to decipher the complexity of epigenetic remodeling which has been linked to the tremendous cellular plasticity seen in PDAC [69,70,71,72]. In the following, we will discuss the impact of epigenetic modifiers that target the lysine residues of histones.
2.1 Histone demethylases and histone methyltransferases
H3K27me3 is commonly linked to transcriptional repression whereas H3K4me3 is typically located at actively transcribed genes [75,76,77]. Active enhancer regions, on the other hand, are generally occupied by monomethylated H3K4 (H3K4me1) [78]. The most frequently mutated histone modifiers in PDAC are KDM6A as well as KMT2D (MLL2) and KMT2C (MLL3), which contribute to the H3K4 and H3K27 methylation landscape [6, 67, 79]. KDM6A, a Jumonji C domain demethylase also known as ubiquitously-transcribed X chromosome tetratricopeptide repeat protein (UTX), can orchestrate the demethylation of H3K27me3 with the destabilization of heterochromatin [80]. In pancreatic acinar cells, KDM6A acts in concert with the homeodomain transcription factor HNF1A to maintain the specific cell identity program [72]. In poorly differentiated cancer entities and PDAC metastasis, KDM6A displays a reduced expression [38, 67]. Moreover, it has been observed that Kdm6a loss accelerates tumor progression and metastasis development in genetically engineered PDAC mouse models, particularly in a gender-specific fashion [38, 67]. This phenomenon is attributed to the functional compensation by the Y chromosome-encoded UTY, which is a member of the KDM6 family but lacks demethylase activity. Mechanistically, the loss of KDM6A causes a demethylase-independent rewiring of the super-enhancer (SE) landscape, resulting in the enforced expression of drivers of dedifferentiation and metastasis, such as ΔNp63, MYC, RUNX3, or ZEB1 [38, 67]. The complex of proteins associated with set1 (COMPASS)-like complex contains the core methyltransferases KMT2C/D and KDM6A [81]. It was shown that the loss of KDM6A leads to an increase in the KMT2D and H3K4me1 signals at SE, which subsequently led to the proposal of a COMPASS-localization function that explains the rewiring of SE [38, 67]. In addition, activin A, a TGF-beta superfamily member, was shown to contribute to EMT via a non-canonical pathway involving the p38 mitogen-activated protein kinase in KDM6A-deficient cells [67].
Importantly, KDM6A-deficient PDACs expose specific therapeutic vulnerabilities. Elegant studies have observed that KDM6A instructs the tumor microenvironment (TME) of PDAC, whereby Kdm6a-deficient murine PDAC cells exhibit increased levels of the CXC motif chemokine ligand 1 (CXCL1) with subsequent recruitment of neutrophils, thus highlighting the CXCL1-CXCR2 axis as a potential therapeutic target [82]. From a tumor cell-intrinsic perspective, KDM6A is involved in regulating the mTORC1 signaling pathway in liver cancers and PDACs, presenting opportunities for the precise use of mTOR inhibitors [83]. Moreover, the loss of the KDM6A demethylase can be exploited using inhibitors of the BET family of proteins [38], as well as histone deacetylase inhibitors, as shown by previous studies [84].
On the opposite end of the H3K27 methylation spectrum, the histone methyltransferase EZH2 installs the repressive H3K27me3 mark. EZH2 is a member of the polycomb group (PcG) proteins, which are chromatin regulators organized in two complexes that are involved in gene silencing. Polycomb repressive complex 1 (PRC1) exposes E3 ubiquitin ligase activity to Lys118 and Lys119 of histone H2A (H2AK118ub and H2AK119ub), while the core polycomb repressive complex 2 (PRC2) contains EED, SUZ12, EZH2 (or EZH1), RBBP4, or RBBP7 [85]. The function of EZH2 is highly context-dependent, as exemplified in the carcinogenesis of the pancreas. In genetically engineered mouse models of PDAC driven by KrasG12D, the genetic inactivation of EZH2 accelerates early carcinogenesis due to a highly inflammatory microenvironment [86]. In contrast to the acceleration observed for early carcinogenesis, the inactivation of one allele of EZH2 results in deaccelerated PDAC development and impaired metastasis formation [70], underscoring the context- and stage-dependent functions of EZH2. One explanation for the context dependency is the crosstalk of EZH2 with the pro-inflammatory and pro-oncogenic transcription factor NFATc1. After a pancreatic injury, EZH2 represses the NFATc1 gene, whereas, in established PDACs, EZH2 maintains NFATc1 expression [39]. Furthermore, the EZH2-dependent dedifferentiation was shown to be mediated by the transcriptional repression of transcription factors GATA6 [70], which is a gatekeeper for epithelial differentiation [87]. Other contextual frameworks determining the functionality of EZH2 and relevant for the efficacy of EZH2 targeting, such as the mutational status of the tumor suppressor p53, were also shown to be important in PDAC [88]. In addition, recent data demonstrate how the methyltransferase cross-talks with the tumor microenvironment in established PDACs. EZH2 represses a pro-inflammatory transcriptome, contributing to profound immune suppression, with impaired NK and T cell surveillance, characteristics for PDAC [89].
The histone methyltransferases KMT2D and KMT2C regulate genomic regions by H3K4-monomethylation (H3K4me1) which is predominantly found at active enhancers co-marked with H3K27ac. These epigenetic genes are frequently mutated in PDACs. In a 2016 study using a PDAC mRNA expression dataset provided by the International Cancer Genome Consortium (ICGC), it was found that pancreatic cancer patients with low KMT2D and KMT2C mRNA expression had better survival rates [90]. Furthermore, a siRNA-dependent knockdown of KMT2C or KMT2D in human PDAC cell lines showed a proliferation-promoting role for both KMTs [90]. Opposingly, increased growth of PDAC cells was observed upon KMT2D knockdown in vitro and in vivo, with reduced expression of KMT2D being linked to metabolic rewiring involving an increased usage of aerobic glycolysis and polyunsaturated fatty acids (PUFAs) to satisfy the increased demands associated with proliferation [91]. Furthermore, the TGF-beta signaling pathway controls KMT2D expression, with metastatic PDAC revealing the lowest KMT2D protein expression [92]. This combined with the connection to the EMT regulator TGF-beta suggests a role for KMT2D in the metastatic cascade. Consistently, CRISPR/Cas9 knock-out of KMT2D in PDAC cell lines led to the upregulation of metastasis-promoting pathways such as MYC, EMT, or angiogenesis [92]. It was observed that KMT2D knock-out cells increase the expression of activin A, which drives EMT via non-canonical p38 MAPK signaling [92]. Moreover, the KMT2D-dependent instruction of the tumor microenvironment might be relevant in PDAC [93], suggesting that epigenetic therapies can tune the tumor cell-intrinsic to -extrinsic rheostat.
2.2 Histone acetyltransferases
The epigenetic writer’s histone acetyltransferases (HATs) are responsible for promoting the accessibility of genes and genomic regions by histone acetylation and subsequent euchromatin formation. Some HATs display additional succinyl-transferase abilities and provide secondary regulatory mechanisms [94,95,96,97]. HATs can be grouped into the families of GNATs, MYST, as well as p300/CBP [98]. The most prominent HAT, E1A binding protein p300, has been well described due to its involvement in the regulation of global transcriptional regulation and its potential to target all histone subunits [99]. In pancreatic cancer, both tumor-promoting and tumor-suppressing functions have been described for p300. The occurrence of the EP300 gene mutation in pancreatic cancer (Fig. 2b) is a hint towards its tumor-suppressive function. On the other hand, multiple studies described an increased activity of the proto-oncogene MYC mediated by p300 in other tumor entities [100, 101]. In a recent study, it was found that mutations in EP300 lead to resistance against inhibitors of porcupine (PORCN), an endoplasmic reticulum O-acyltransferase, in RNF43-mutant pancreatic cancers [102]. PORCN is responsible for the palmitoylation of Wnts, which is necessary for their secretion and activation of the receptors [103, 104]. Approximately 5–10% of PDAC cases contain mutations in the RNF43 E3 ubiquitin ligase [6, 105], which can direct Wnt surface receptors to the proteasome [106, 107]. Therefore, PDACs with inactivated RNF43 express a higher level of Wnt receptors, such as Frizzleds (FZDs) and LRP5/6, making them more susceptible to inhibitors targeting ligand-activated Wnt signaling [108]. p300 plays a crucial role in the expression of GATA6, and the inactivation of EP300 leads to dedifferentiation and a shift towards a more basal-like subtype. This dedifferentiation process, which bypasses WNT-dependent pathways, explains the resistance of RNF43/EP300 double mutant PDACs to PORCN inhibitors [102].
Furthermore, elevated levels of miRNAs that target p300 and subsequent low expression of p300 were observed in PDAC cell lines displaying high metastatic potential in orthotropic mouse models [109]. Irrespective of metastatic potential, multiple studies have shown that inhibition of p300 and its coactivator CBP also improved gemcitabine sensitivity and reduced cell-cycle progression and proliferation in PDAC cells [110,111,112,113]. Shi et al. elaborated on this topic and observed a correlation between platelet-derived growth factor C (PDGFC) expression and gemcitabine resistance as a result of platelet-derived growth factor receptor alpha (PDGFRα) and beta (PDGFRβ) activation. They showed that p300 binds to the promoter site of the PDGFC gene, and inhibition of p300 reduced PDGFC expression [113]. Since PDGFRβ signaling has been linked to a more aggressive and metastatic phenotype in PDAC [114, 115], the observed regulation of PDGFC by p300 provides evidence for a more aggressive phenotype orchestrated by p300-dependent chromatin remodeling [113]. In a recent study, promising findings in mice regarding a novel dual inhibitor, XP-524, targeting BET proteins and p300 were presented. The results demonstrated improved survival and a reorganized microenvironment upon XP-524 treatment, resulting in enhanced immune infiltration [116]. Furthermore, the researchers noted that the inclusion of an anti-PD-1 antibody in the treatment regimen further enhanced survival. These findings suggest the potential of XP-524 as a therapeutic approach, possibly in combination with immune checkpoint inhibitors, for improved outcomes in the future [116]. Regarding other HATs, researchers discovered an increased expression of histone acetyltransferase 1 (HAT1) in pancreatic tumor tissue compared to healthy tissue, and high expression of HAT1 correlated with a worse prognosis [117]. The knockdown of HAT1 resulted in reduced proliferation and tumor volume in the investigated human cell lines and murine models. The researchers additionally observed a correlation between HAT1 and PD-L1 expression and identified increased recruitment of BRD4 to the PD-L1 promoter in a HAT1-dependent manner. Whether HAT1 mediates BRD4 recruitment via H4K5 acetylation or its newly discovered succinyl-transferase activity remains unclear [94, 117]. Regardless, the results indicate that HAT1 might contribute to immune evasion during PDAC progression. Similar to HAT1, lysine acetyltransferase 2A (KAT2A/GCN5) also showed increased, aberrant expression in PDAC tissue and significantly worse overall survival in the KAT2Ahigh group in a cohort of 40 patients [97]. KAT2A knockdown resulted in reduced proliferation, impaired wound healing, and decreased invasion ability. Mechanistically, KAT2A was found to transcriptionally regulate YWHAZ (also known as 14-3-3ζ) through H3K79 succinylation [97]. 14-3-3ζ stabilizes β-catenin and maintains canonical Wnt signaling [118]. The knockdown of KAT2A led to reduced expression of 14-3-3ζ and subsequent degradation of β-catenin [97]. This, in turn, resulted in decreased expression of β-catenin target genes, including c-MYC, GLUT1, LDHA, and cyclin D1. Interestingly, the researchers attempted to restore the phenotype of KAT2A knockdown cells by reintroducing KAT2Awt. They observed successful reversion of the phenotype; however, when they reconstituted a succinyl-transferase defective form of KAT2A (KAT2AY645A), they failed to restore the expression of 14-3-3ζ and H3K79 succinylation, despite the rescue of the H3K9 acetylation. These results emphasize the significance of acetylation-independent functions of KAT2A, particularly concerning the aggressiveness of PDAC [97].
Finally, Ono et al. analyzed immunohistochemistry staining of H3K9 and H3K27 acetylation in a cohort of 102 pancreatic cancer patients indicating a significantly worse overall survival in patients with intermediate staining compared to patients with high/low staining [111]. Considering that changes in H3K27ac precede enhancer reprogramming and are essential for cellular plasticity, it is not surprising that HATs may possess dual-functional roles in PDAC. The results of Ono et al. imply that preservation of a plastic acetylation state might lead to the worst prognosis and resembles the concept of the previously described p-EMT phenotype.
2.3 Histone deacetylases
The cellular counterparts to HATs are histone deacetylases (HDACs). The family of HDACs has been well conserved across evolution. They are generally grouped into four different classes according to the sequence homologies with their corresponding yeast homolog [119, 120]. Classes I, II, and IV contain a Zn2+-ion in their enzymatic center whereas members of class III, also called sirtuins, are reliant on NAD+ as a cofactor and are therefore often regarded separately [121]. HDAC1-3 as well as HDAC8, belong to class I whereas HDAC4, 5, and 7 belong to class IIa, and HDAC6 and HDAC10 belong to class IIb respectively. HDAC11 is the sole identified member of class IV (Fig. 3a). HDACs primarily control gene transcription by deacetylation of the lysine residues at the histone tails causing condensation of the chromatin at the deacetylated regions which in turn prevents the binding of the transcriptional machinery [119, 122, 123]. The recruitment of HDACs to their designated target region on the genome is mainly coordinated by different repressor complexes. Those complexes include proteins with histone-recognition motifs like inhibitor of growth family member 2 (ING2) which recognizes and binds to H3K4me3 and can interact with transcription factors that require HDAC recruitment for their repressive functions [124, 125].
Histone deacetylases in PDAC. a Phylogenic tree of human HDACs. b Pearson correlation between metastatic/penetrance potential and RNA expression of class I HDACs and repressor complexes. Positive correlation is depicted in green and negative correlation is depicted in purple. *, P<0.05; **, P<0.01; ***, P<0.001. Amino acid sequences of human HDACs were downloaded from uniprot [136], and phylogenic tree was generated using simple phylogeny [137, 138] and ggtree [139]. Correlation coefficient data was downloaded from DepMap [140,141,142]
Although mutations of HDACs are not commonly found in PDAC, several studies have reported increased expression of HDACs in tumor tissue compared to normal pancreatic tissue [126,127,128]. Among the HDACs, class I HDACs, particularly HDAC1, HDAC2, and HDAC3, are highly represented [128]. Furthermore, previous studies have indicated that less differentiated tumors exhibit elevated expression of class I HDACs [127, 129]. A closer examination of the metastasis map [99,100,101], whose results are based on the implantation of cancer cell lines into mice and allows for the association of metastasis patterns with clinical and genomic characteristics, has confirmed a link between class I HDACs and repressor complexes with PDAC metastasis (Fig. 3b). While HDAC1 showed a correlation with overall metastatic potential in the 30 available PDAC cell lines, HDAC2 and HDAC8 expression was particularly associated with organ-specific metastasis to the liver. HDAC3 did not directly correlate with metastatic potential. However, the subunits of the major repressor complexes of class I HDACs, specifically the corepressor subunits RCOR2 (COREST2), NCOR1, and NCOR2 (SMRT), exhibited a strong correlation with overall metastasis potential in the PDAC metastasis map (Fig. 3b). NCOR1 and NCOR2 are mainly responsible for recruiting HDAC3 to facilitate histone deacetylation [130], while COREST recruits HDAC1 and HDAC2 [131].
The findings regarding the deacetylase-controlled metastatic potential align with a recently published study conducted on genetically defined murine models of autochthonous PDAC. The study revealed a significant reduction in metastasis formation in Hdac2-deficient murine PDACs [69]. In Hdac2-deficient cells, the expression of several tyrosine kinase receptors (RTKs), including PDGFRα and PDGFRβ, was found to be decreased [69]. RTKs, including the ones mentioned, likely play a crucial role in ensuring cell survival throughout the different stages of the metastatic cascade [114, 115]. Moreover, a correlation between HDAC2 and the TGF-beta signaling pathway was observed. In the absence of HDAC2, murine PDAC cells undergoing TGF-beta-driven EMT exhibited a significant increase in reactive oxygen species (ROS), ultimately leading to cell death. These findings suggest that HDAC2 plays a role in orchestrating a program to detoxify ROS and in limiting the tumor-suppressive effects of TGF-beta [69]. Furthermore, TGF-beta downstream regulation was partially impaired in Hdac2-deficient cells as evidenced by impaired repression of the epithelial marker E-cadherin (Cdh1) and SNAI1 upregulation [69], which is concordant with earlier reports [132] and observations that HDAC inhibitor (HDACi) Domatinostat (4SC-202) attenuated TGF-beta signaling, including reduced expression of the EMT transcription factor ZEB1, as well as induced upregulation of genes in a BRD4 and MYC-dependent manner [133].
Additional experimental evidence has highlighted the role of HDAC1 in PDAC metastasis. Treatment of PDAC cells with HDACis, such as DHOB and vorinostat, resulted in a reduction in their invasive potential. This was accompanied by a decrease in the expression of the EMT transcription factor SNAI1 [129]. Furthermore, a cohort of 103 PDAC patients categorized based on HDAC1 protein expression revealed that patients with high expression of this class I deacetylase exhibited significantly lower distant metastasis-free survival (DMFS) [129].
Supporting these results, in a pooled CRISPR screen in human PDAC cells, the correlation between HDAC1 overexpression and the induction of EMT genes, along with an increase in cell migration was described [134].
In summary, there is a clear association between HDAC1 and HDAC2 and the metastasis of PDAC, presenting an opportunity for therapeutic intervention. However, it should be noted that a study in melanoma cells observed an epigenetic silencing of invasiveness genes partly via HDAC2 [135]. Delineation of such contextual differences will increase the understanding of how HDACs direct metastasis.
3 Development of epigenetic PDAC therapies
With advances in drug development and improved methods of on-target detection, the opportunities for novel-targeted treatments are vastly growing. Proteomic assays confirming target specificity have recently shown surprising results in HDACi specificity and show remarkable differences concerning HDACi potency when targeting HDACs within their designated repressor complexes [143].
To address target specificity and target accessibility, drugs with new mechanisms of action are on the rise. Protein degraders such as PROTACs might offer opportunities to target proteins more specifically than prior and will allow addressing functions independent of the enzymatic activity [144, 145]. A promising example in PDAC treatment is the progress made with KRAS mutant-specific degraders [145]. In parallel, efforts to develop novel degraders for epigenetic targets are also increasing [144]. So far, most clinically approved epigenetic drugs have found applications in non-solid tumors, such as DNA methyltransferase (DNMT) inhibitors 5-azacytidine and 5-Aza-2-deoxycytidine for the treatment of myelodysplastic syndrome [146, 147]. The HDACis vorinostat, romidepsin, and belinostat have been approved for the treatment of refractory cutaneous T cell lymphoma and refractory peripheral T cell lymphoma, respectively [148, 149]. Another HDACi, panobinostat, which had been approved for the treatment of multiple myeloma in 2015, has since been revoked by the FDA on March 2022 due to a lack of follow-up data, questioning the value of HDACis in the clinic. In 2020, however, the EZH2-inhibitor tazemetostat was approved for the treatment of advanced epithelioid sarcomas, as the first epigenetic drug with an application in solid tumors [150], underscoring the potential of epigenetic therapies.
In PDAC, epigenetic drugs have yet to be approved. Considering the potential benefits that have been described in preclinical studies, current clinical studies have shown increasing interest. Recent efforts observed that priming primary PDAC cells with a selection of epigenetic drugs sensitized the tested cell line to chemotherapeutic treatment [151]. However, the overall response across cell lines displayed a high heterogeneity concerning the individual drugs. To predict beneficial priming and chemotherapeutic therapies, the researchers generated transcriptomic predictor signatures [151]. This study exemplifies the rationale for epigenetic-chemotherapeutic combination therapies and their stratification strategies. Additionally, at present, there are a variety of studies ongoing or in the process of patient recruitment that focus on the treatment of PDAC patients with epigenetic drugs (Table 1). A phase-II clinical trial including the previously mentioned tazemetostat is currently being initiated and combines the EZH2 inhibitor with the immune therapeutic durvalumab (NCT04705818). This clinical trial might yield promising results, as recent studies connected EZH2 to an immune suppressive microenvironment [89]. DNMT inhibitor azacitidine is also being investigated in multiple studies that include immune checkpoint inhibitors and chemotherapy (NCT03264404, NCT04257448). DNMTs were shown to contribute to PDAC metastasis and their inhibition in other solid tumors-potentiated anti-tumor immune responses [152]. Another trial involving azacitidine has recently been completed that investigates tumor recurrence after tumor resection and adjuvant treatment (NCT01845805). Unfortunately, the study examiners did not observe a significant difference in relapse or overall survival between adjuvant azacitidine treatment or control groups that did not receive any additional treatment [153].
Despite showing little to no success in solid tumors so far, a variety of HDACis have also been included in recent combination studies with other treatment regimens including BET inhibitors (NCT05053971), as well as checkpoint inhibitors (NCT03250273, NCT04257448) and classical chemotherapy (NCT02349867, NCT03878524, NCT02737228). These studies include both class I-specific HDACis such as entinostat and romidepsin but also inhibitors with a broader target range such as panobinostat and vorinostat as well as ivaltinostat. Another clinical trial included vorinostat treatment to the neoadjuvant radiotherapy regimen based on previous sensitization results (NCT02349867) [154]. The recently finished phase I/II trials combining ivaltinostat with gemcitabine and erlotinib treatment showed promising results regarding toxicity and patient survival. However, due to its single-arm design, results must be further validated (NCT02737228) [155]. The common side effects of epigenetic therapies are illustrated in Table 2. However, concepts to lower adverse effects, like sequential scheduling or using low doses below the maximally tolerated dose maintain efficacy in pre-clinical models exist [156,157,158] and therefore, such concepts offer clinical opportunities to reduce adverse effects.
4 Conclusion
As research technology continues to advance, therapeutic stratification strategies for cancer patients are undergoing significant changes. The advancements in multi-omics analysis are opening the door to a new era of treatment regimens and improved targeting of specific subgroups. However, the complex nature of cancer, including cellular plasticity, mainly controlled by transcriptional and epigenetic flexibility, remains challenging.
Previous studies have demonstrated that metastasizing cells rely heavily on their adaptive capabilities. As they progress through the metastatic cascade, they must quickly overcome various challenges, including starvation, migration, invasion, and survival in the bloodstream, before establishing themselves at a distant organ site. Therefore, it comes as no surprise that disseminating cells heavily depend on epigenetic regulators. However, it is important to note that chromatin dynamics are highly complex and only partially understood. Additionally, emerging research suggests that histone-modifying enzymes not only modify histones but also alter the functionality of other proteins. Consequently, gaining a more comprehensive understanding of the role of epigenetic regulators in tumor progression will be crucial for the successful and precise design of clinical studies.
References
Siegel, R. L., Miller, K. D., Wagle, N. S., & Jemal, A. (2023). Cancer statistics, 2023. CA: a Cancer Journal for Clinicians, 73(1), 17–48. https://doi.org/10.3322/caac.21763
Park, W., Chawla, A., & O'Reilly, E. M. (2021). Pancreatic cancer: A review. JAMA, 326(9), 851–862. https://doi.org/10.1001/jama.2021.13027
Halbrook, C. J., Lyssiotis, C. A., Pasca di Magliano, M., & Maitra, A. (2023). Pancreatic cancer: Advances and challenges. Cell, 186(8), 1729–1754. https://doi.org/10.1016/j.cell.2023.02.014
Rhim, A. D., Mirek, E. T., Aiello, N. M., Maitra, A., Bailey, J. M., McAllister, F., Reichert, M., Beatty, G. L., Rustgi, A. K., Vonderheide, R. H., Leach, S. D., & Stanger, B. Z. (2012). EMT and dissemination precede pancreatic tumor formation. Cell, 148(1-2), 349–361. https://doi.org/10.1016/j.cell.2011.11.025
Haeno, H., Gonen, M., Davis, M. B., Herman, J. M., Iacobuzio-Donahue, C. A., & Michor, F. (2012). Computational modeling of pancreatic cancer reveals kinetics of metastasis suggesting optimum treatment strategies. Cell, 148(1-2), 362–375. https://doi.org/10.1016/j.cell.2011.11.060
Waddell, N., Pajic, M., Patch, A. M., Chang, D. K., Kassahn, K. S., Bailey, P., Johns, A. L., Miller, D., Nones, K., Quek, K., Quinn, M. C., Robertson, A. J., Fadlullah, M. Z., Bruxner, T. J., Christ, A. N., Harliwong, I., Idrisoglu, S., Manning, S., Nourse, C., & Grimmond, S. M. (2015). Whole genomes redefine the mutational landscape of pancreatic cancer. Nature, 518(7540), 495–501. https://doi.org/10.1038/nature14169
Nguyen, B., Fong, C., Luthra, A., Smith, S. A., DiNatale, R. G., Nandakumar, S., Walch, H., Chatila, W. K., Madupuri, R., Kundra, R., Bielski, C. M., Mastrogiacomo, B., Donoghue, M. T. A., Boire, A., Chandarlapaty, S., Ganesh, K., Harding, J. J., Iacobuzio-Donahue, C. A., Razavi, P., & Schultz, N. (2022). Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell, 185(3), 563–575 e511. https://doi.org/10.1016/j.cell.2022.01.003
Connor, A. A., Denroche, R. E., Jang, G. H., Lemire, M., Zhang, A., Chan-Seng-Yue, M., Wilson, G., Grant, R. C., Merico, D., Lungu, I., Bartlett, J. M. S., Chadwick, D., Liang, S. B., Eagles, J., Mbabaali, F., Miller, J. K., Krzyzanowski, P., Armstrong, H., Luo, X., & Gallinger, S. (2019). Integration of genomic and transcriptional features in pancreatic cancer reveals increased cell cycle progression in metastases. Cancer Cell, 35(2), 267–282 e267. https://doi.org/10.1016/j.ccell.2018.12.010
Makohon-Moore, A. P., Zhang, M., Reiter, J. G., Bozic, I., Allen, B., Kundu, D., Chatterjee, K., Wong, F., Jiao, Y., Kohutek, Z. A., Hong, J., Attiyeh, M., Javier, B., Wood, L. D., Hruban, R. H., Nowak, M. A., Papadopoulos, N., Kinzler, K. W., Vogelstein, B., & Iacobuzio-Donahue, C. A. (2017). Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nature Genetics, 49(3), 358–366. https://doi.org/10.1038/ng.3764
McDonald, O. G., Li, X., Saunders, T., Tryggvadottir, R., Mentch, S. J., Warmoes, M. O., Word, A. E., Carrer, A., Salz, T. H., Natsume, S., Stauffer, K. M., Makohon-Moore, A., Zhong, Y., Wu, H., Wellen, K. E., Locasale, J. W., Iacobuzio-Donahue, C. A., & Feinberg, A. P. (2017). Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nature Genetics, 49(3), 367–376. https://doi.org/10.1038/ng.3753
Mueller, S., Engleitner, T., Maresch, R., Zukowska, M., Lange, S., Kaltenbacher, T., Konukiewitz, B., Ollinger, R., Zwiebel, M., Strong, A., Yen, H. Y., Banerjee, R., Louzada, S., Fu, B., Seidler, B., Gotzfried, J., Schuck, K., Hassan, Z., Arbeiter, A., & Rad, R. (2018). Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature, 554(7690), 62–68. https://doi.org/10.1038/nature25459
Chan-Seng-Yue, M., Kim, J. C., Wilson, G. W., Ng, K., Figueroa, E. F., O'Kane, G. M., Connor, A. A., Denroche, R. E., Grant, R. C., McLeod, J., Wilson, J. M., Jang, G. H., Zhang, A., Dodd, A., Liang, S. B., Borgida, A., Chadwick, D., Kalimuthu, S., Lungu, I., & Notta, F. (2020). Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nature Genetics, 52(2), 231–240. https://doi.org/10.1038/s41588-019-0566-9
Maddipati, R., Norgard, R. J., Baslan, T., Rathi, K. S., Zhang, A., Saeid, A., Higashihara, T., Wu, F., Kumar, A., Annamalai, V., Bhattacharya, S., Raman, P., Adkisson, C. A., Pitarresi, J. R., Wengyn, M. D., Yamazoe, T., Li, J., Balli, D., LaRiviere, M. J., & Stanger, B. Z. (2022). MYC levels regulate metastatic heterogeneity in pancreatic adenocarcinoma. Cancer Discovery, 12(2), 542–561. https://doi.org/10.1158/2159-8290.CD-20-1826
Brar, G., Blais, E. M., Joseph Bender, R., Brody, J. R., Sohal, D., Madhavan, S., Picozzi, V. J., Hendifar, A. E., Chung, V. M., Halverson, D., Mikhail, S., Matrisian, L. M., Rahib, L., Petricoin, E., & Pishvaian, M. J. (2019). Multi-omic molecular comparison of primary versus metastatic pancreatic tumours. British Journal of Cancer, 121(3), 264–270. https://doi.org/10.1038/s41416-019-0507-5
Burdziak, C., Alonso-Curbelo, D., Walle, T., Reyes, J., Barriga, F. M., Haviv, D., Xie, Y., Zhao, Z., Zhao, C. J., Chen, H. A., Chaudhary, O., Masilionis, I., Choo, Z. N., Gao, V., Luan, W., Wuest, A., Ho, Y. J., Wei, Y., Quail, D. F., & Pe’er, D. (2023). Epigenetic plasticity cooperates with cell-cell interactions to direct pancreatic tumorigenesis. Science, 380(6645), eadd5327. https://doi.org/10.1126/science.add5327
Roe, J. S., Hwang, C. I., Somerville, T. D. D., Milazzo, J. P., Lee, E. J., Da Silva, B., Maiorino, L., Tiriac, H., Young, C. M., Miyabayashi, K., Filippini, D., Creighton, B., Burkhart, R. A., Buscaglia, J. M., Kim, E. J., Grem, J. L., Lazenby, A. J., Grunkemeyer, J. A., Hollingsworth, M. A., & Vakoc, C. R. (2017). Enhancer reprogramming promotes pancreatic cancer metastasis. Cell, 170(5), 875–888 e820. https://doi.org/10.1016/j.cell.2017.07.007
Milan, M., Balestrieri, C., Alfarano, G., Polletti, S., Prosperini, E., Spaggiari, P., Zerbi, A., Diaferia, G. R., & Natoli, G. (2019). FOXA2 controls the cis-regulatory networks of pancreatic cancer cells in a differentiation grade-specific manner. The EMBO Journal, 38(20), e102161. https://doi.org/10.15252/embj.2019102161
Chiou, S. H., Risca, V. I., Wang, G. X., Yang, D., Gruner, B. M., Kathiria, A. S., Ma, R. K., Vaka, D., Chu, P., Kozak, M., Castellini, L., Graves, E. E., Kim, G. E., Mourrain, P., Koong, A. C., Giaccia, A. J., & Winslow, M. M. (2017). BLIMP1 induces transient metastatic heterogeneity in pancreatic cancer. Cancer Discovery, 7(10), 1184–1199. https://doi.org/10.1158/2159-8290.CD-17-0250
Whittle, M. C., Izeradjene, K., Rani, P. G., Feng, L., Carlson, M. A., DelGiorno, K. E., Wood, L. D., Goggins, M., Hruban, R. H., Chang, A. E., Calses, P., Thorsen, S. M., & Hingorani, S. R. (2015). RUNX3 controls a metastatic switch in pancreatic ductal adenocarcinoma. Cell, 161(6), 1345–1360. https://doi.org/10.1016/j.cell.2015.04.048
Krebs, A. M., Mitschke, J., Lasierra Losada, M., Schmalhofer, O., Boerries, M., Busch, H., Boettcher, M., Mougiakakos, D., Reichardt, W., Bronsert, P., Brunton, V. G., Pilarsky, C., Winkler, T. H., Brabletz, S., Stemmler, M. P., & Brabletz, T. (2017). The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nature Cell Biology, 19(5), 518–529. https://doi.org/10.1038/ncb3513
Takano, S., Reichert, M., Bakir, B., Das, K. K., Nishida, T., Miyazaki, M., Heeg, S., Collins, M. A., Marchand, B., Hicks, P. D., Maitra, A., & Rustgi, A. K. (2016). Prrx1 isoform switching regulates pancreatic cancer invasion and metastatic colonization. Genes & Development, 30(2), 233–247. https://doi.org/10.1101/gad.263327.115
Dai, C., Rennhack, J. P., Arnoff, T. E., Thaker, M., Younger, S. T., Doench, J. G., Huang, A. Y., Yang, A., Aguirre, A. J., Wang, B., Mun, E., O'Connell, J. T., Huang, Y., Labella, K., Talamas, J. A., Li, J., Ilic, N., Hwang, J., Hong, A. L., & Hahn, W. C. (2021). SMAD4 represses FOSL1 expression and pancreatic cancer metastatic colonization. Cell Reports, 36(4), 109443. https://doi.org/10.1016/j.celrep.2021.109443
Schneeweis, C., Diersch, S., Hassan, Z., Krauss, L., Schneider, C., Lucarelli, D., Falcomata, C., Steiger, K., Ollinger, R., Kramer, O. H., Arlt, A., Grade, M., Schmidt-Supprian, M., Hessmann, E., Wirth, M., Rad, R., Reichert, M., Saur, D., & Schneider, G. (2022). AP1/Fra1 confers resistance to MAPK cascade inhibition in pancreatic cancer. Cellular and Molecular Life Sciences, 80(1), 12. https://doi.org/10.1007/s00018-022-04638-y
Hamdan, F. H., & Johnsen, S. A. (2018). DeltaNp63-dependent super enhancers define molecular identity in pancreatic cancer by an interconnected transcription factor network. Proceedings of the National Academy of Sciences, 115(52), E12343–E12352. https://doi.org/10.1073/pnas.1812915116
Somerville, T. D. D., Xu, Y., Miyabayashi, K., Tiriac, H., Cleary, C. R., Maia-Silva, D., Milazzo, J. P., Tuveson, D. A., & Vakoc, C. R. (2018). TP63-mediated enhancer reprogramming drives the squamous subtype of pancreatic ductal adenocarcinoma. Cell Reports, 25(7), 1741–1755 e1747. https://doi.org/10.1016/j.celrep.2018.10.051
Sahin, I. H., Elias, H., Chou, J. F., Capanu, M., & O’Reilly, E. M. (2018). Pancreatic adenocarcinoma: Insights into patterns of recurrence and disease behavior. BMC Cancer, 18(1), 769. https://doi.org/10.1186/s12885-018-4679-9
Yachida, S., White, C. M., Naito, Y., Zhong, Y., Brosnan, J. A., Macgregor-Das, A. M., Morgan, R. A., Saunders, T., Laheru, D. A., Herman, J. M., Hruban, R. H., Klein, A. P., Jones, S., Velculescu, V., Wolfgang, C. L., & Iacobuzio-Donahue, C. A. (2012). Clinical significance of the genetic landscape of pancreatic cancer and implications for identification of potential long-term survivors. Clinical Cancer Research, 18(22), 6339–6347. https://doi.org/10.1158/1078-0432.CCR-12-1215
Carstens, J. L., Yang, S., Correa de Sampaio, P., Zheng, X., Barua, S., McAndrews, K. M., Rao, A., Burks, J. K., Rhim, A. D., & Kalluri, R. (2021). Stabilized epithelial phenotype of cancer cells in primary tumors leads to increased colonization of liver metastasis in pancreatic cancer. Cell Reports, 35(2), 108990. https://doi.org/10.1016/j.celrep.2021.108990
Krauss, L., Urban, B. C., Hastreiter, S., Schneider, C., Wenzel, P., Hassan, Z., Wirth, M., Lankes, K., Terrasi, A., Klement, C., Cernilogar, F. M., Ollinger, R., de Andrade Kratzig, N., Engleitner, T., Schmid, R. M., Steiger, K., Rad, R., Kramer, O. H., Reichert, M., & Schneider, G. (2022). HDAC2 facilitates pancreatic cancer metastasis. Cancer Research, 82(4), 695–707. https://doi.org/10.1158/0008-5472.CAN-20-3209
Cheng, P., Chen, Y., He, T. L., Wang, C., Guo, S. W., Hu, H., Ni, C. M., Jin, G., & Zhang, Y. J. (2019). Menin coordinates C/EBPbeta-mediated TGF-beta signaling for epithelial-mesenchymal transition and growth inhibition in pancreatic cancer. Molecular Therapy--Nucleic Acids, 18, 155–165. https://doi.org/10.1016/j.omtn.2019.08.013
Chang, C. J., Chao, C. H., Xia, W., Yang, J. Y., Xiong, Y., Li, C. W., Yu, W. H., Rehman, S. K., Hsu, J. L., Lee, H. H., Liu, M., Chen, C. T., Yu, D., & Hung, M. C. (2011). p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nature Cell Biology, 13(3), 317–323. https://doi.org/10.1038/ncb2173
Aiello, N. M., Maddipati, R., Norgard, R. J., Balli, D., Li, J., Yuan, S., Yamazoe, T., Black, T., Sahmoud, A., Furth, E. E., Bar-Sagi, D., & Stanger, B. Z. (2018). EMT subtype influences epithelial plasticity and mode of cell migration. Developmental Cell, 45(6), 681–695 e684. https://doi.org/10.1016/j.devcel.2018.05.027
Semaan, A., Bernard, V., Kim, D. U., Lee, J. J., Huang, J., Kamyabi, N., Stephens, B. M., Qiao, W., Varadhachary, G. R., Katz, M. H., Shen, Y., San Lucas, F. A., Gascoyne, P., Alvarez, H. A., Maitra, A., & Guerrero, P. A. (2021). Characterisation of circulating tumour cell phenotypes identifies a partial-EMT sub-population for clinical stratification of pancreatic cancer. British Journal of Cancer, 124(12), 1970–1977. https://doi.org/10.1038/s41416-021-01350-9
Yuan, S., Norgard, R. J., & Stanger, B. Z. (2019). Cellular plasticity in cancer. Cancer Discovery, 9(7), 837–851. https://doi.org/10.1158/2159-8290.CD-19-0015
Gerstberger, S., Jiang, Q., & Ganesh, K. (2023). Metastasis. Cell, 186(8), 1564–1579. https://doi.org/10.1016/j.cell.2023.03.003
van Roey, R., Brabletz, T., Stemmler, M. P., & Armstark, I. (2021). Deregulation of transcription factor networks driving cell plasticity and metastasis in pancreatic cancer. Frontiers in Cell and Developmental Biology, 9, 753456. https://doi.org/10.3389/fcell.2021.753456
Lan, L., Evan, T., Li, H., Hussain, A., Ruiz, E. J., Zaw Thin, M., Ferreira, R. M. M., Ps, H., Riising, E. M., Zen, Y., Almagro, J., Ng, K. W., Soro-Barrio, P., Nelson, J., Koifman, G., Carvalho, J., Nye, E. L., He, Y., Zhang, C., & Behrens, A. (2022). GREM1 is required to maintain cellular heterogeneity in pancreatic cancer. Nature, 607(7917), 163–168. https://doi.org/10.1038/s41586-022-04888-7
Andricovich, J., Perkail, S., Kai, Y., Casasanta, N., Peng, W., & Tzatsos, A. (2018). Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell, 33(3), 512–526 e518. https://doi.org/10.1016/j.ccell.2018.02.003
Patil, S., Forster, T., Reutlinger, K., Kopp, W., Versemann, L., Spitalieri, J., Gaedcke, J., Strobel, P., Singh, S. K., Ellenrieder, V., Neesse, A., & Hessmann, E. (2021). Chromatin-independent interplay of NFATc1 and EZH2 in pancreatic cancer. Cells, 10(12). https://doi.org/10.3390/cells10123463
Lomberk, G., Blum, Y., Nicolle, R., Nair, A., Gaonkar, K. S., Marisa, L., Mathison, A., Sun, Z., Yan, H., Elarouci, N., Armenoult, L., Ayadi, M., Ordog, T., Lee, J. H., Oliver, G., Klee, E., Moutardier, V., Gayet, O., Bian, B., & Urrutia, R. (2018). Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nature Communications, 9(1), 1978. https://doi.org/10.1038/s41467-018-04383-6
Massague, J., & Ganesh, K. (2021). Metastasis-initiating cells and ecosystems. Cancer Discovery, 11(4), 971–994. https://doi.org/10.1158/2159-8290.CD-21-0010
Espinet, E., Gu, Z., Imbusch, C. D., Giese, N. A., Buscher, M., Safavi, M., Weisenburger, S., Klein, C., Vogel, V., Falcone, M., Insua-Rodriguez, J., Reitberger, M., Thiel, V., Kossi, S. O., Muckenhuber, A., Sarai, K., Lee, A. Y. L., Backx, E., Zarei, S., & Trumpp, A. (2021). Aggressive PDACs show hypomethylation of repetitive elements and the execution of an intrinsic IFN program linked to a ductal cell of origin. Cancer Discovery, 11(3), 638–659. https://doi.org/10.1158/2159-8290.CD-20-1202
Feinberg, A. P., & Levchenko, A. (2023). Epigenetics as a mediator of plasticity in cancer. Science, 379(6632), eaaw3835. https://doi.org/10.1126/science.aaw3835
Hamdan, F. H., Abdelrahman, A. M., Kutschat, A. P., Wang, X., Ekstrom, T. L., Jalan-Sakrikar, N., Wegner Wippel, C., Taheri, N., Tamon, L., Kopp, W., Aggrey-Fynn, J., Bhagwate, A. V., Alva-Ruiz, R., Lynch, I., Yonkus, J., Kosinsky, R. L., Gaedcke, J., Hahn, S. A., Siveke, J. T., & Johnsen, S. A. (2023). Interactive enhancer hubs (iHUBs) mediate transcriptional reprogramming and adaptive resistance in pancreatic cancer. Gut, 72(6), 1174–1185. https://doi.org/10.1136/gutjnl-2022-328154
Tie, F., Banerjee, R., Saiakhova, A. R., Howard, B., Monteith, K. E., Scacheri, P. C., Cosgrove, M. S., & Harte, P. J. (2014). Trithorax monomethylates histone H3K4 and interacts directly with CBP to promote H3K27 acetylation and antagonize polycomb silencing. Development, 141(5), 1129–1139. https://doi.org/10.1242/dev.102392
Tie, F., Banerjee, R., Stratton, C. A., Prasad-Sinha, J., Stepanik, V., Zlobin, A., Diaz, M. O., Scacheri, P. C., & Harte, P. J. (2009). CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development, 136(18), 3131–3141. https://doi.org/10.1242/dev.037127
Zhang, T., Cooper, S., & Brockdorff, N. (2015). The interplay of histone modifications - writers that read. EMBO Reports, 16(11), 1467–1481. https://doi.org/10.15252/embr.201540945
Lee, J. S., Smith, E., & Shilatifard, A. (2010). The language of histone crosstalk. Cell, 142(5), 682–685. https://doi.org/10.1016/j.cell.2010.08.011
Soares, L. M., He, P. C., Chun, Y., Suh, H., Kim, T., & Buratowski, S. (2017). Determinants of histone H3K4 methylation patterns. Molecular Cell, 68(4), 773–785 e776. https://doi.org/10.1016/j.molcel.2017.10.013
Zhao, W., Xu, Y., Wang, Y., Gao, D., King, J., Xu, Y., & Liang, F. S. (2021). Investigating crosstalk between H3K27 acetylation and H3K4 trimethylation in CRISPR/dCas-based epigenome editing and gene activation. Science Reports, 11(1), 15912. https://doi.org/10.1038/s41598-021-95398-5
Cai, Y., Zhang, Y., Loh, Y. P., Tng, J. Q., Lim, M. C., Cao, Z., Raju, A., Lieberman Aiden, E., Li, S., Manikandan, L., Tergaonkar, V., Tucker-Kellogg, G., & Fullwood, M. J. (2021). H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions. Nature Communications, 12(1), 719. https://doi.org/10.1038/s41467-021-20940-y
Stathis, A., & Bertoni, F. (2018). BET proteins as targets for anticancer treatment. Cancer Discovery, 8(1), 24–36. https://doi.org/10.1158/2159-8290.CD-17-0605
Belkina, A. C., & Denis, G. V. (2012). BET domain co-regulators in obesity, inflammation and cancer. Nature Reviews Cancer, 12(7), 465–477. https://doi.org/10.1038/nrc3256
Winter, G. E., Mayer, A., Buckley, D. L., Erb, M. A., Roderick, J. E., Vittori, S., Reyes, J. M., di Iulio, J., Souza, A., Ott, C. J., Roberts, J. M., Zeid, R., Scott, T. G., Paulk, J., Lachance, K., Olson, C. M., Dastjerdi, S., Bauer, S., Lin, C. Y., & Bradner, J. E. (2017). BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Molecular Cell, 67(1), 5–18 e19. https://doi.org/10.1016/j.molcel.2017.06.004
Jang, M. K., Mochizuki, K., Zhou, M., Jeong, H. S., Brady, J. N., & Ozato, K. (2005). The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Molecular Cell, 19(4), 523–534. https://doi.org/10.1016/j.molcel.2005.06.027
Mazur, P. K., Herner, A., Mello, S. S., Wirth, M., Hausmann, S., Sanchez-Rivera, F. J., Lofgren, S. M., Kuschma, T., Hahn, S. A., Vangala, D., Trajkovic-Arsic, M., Gupta, A., Heid, I., Noel, P. B., Braren, R., Erkan, M., Kleeff, J., Sipos, B., Sayles, L. C., & Siveke, J. T. (2015). Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nature Medicine, 21(10), 1163–1171. https://doi.org/10.1038/nm.3952
Bian, B., Bigonnet, M., Gayet, O., Loncle, C., Maignan, A., Gilabert, M., Moutardier, V., Garcia, S., Turrini, O., Delpero, J. R., Giovannini, M., Grandval, P., Gasmi, M., Ouaissi, M., Secq, V., Poizat, F., Nicolle, R., Blum, Y., Marisa, L., & Iovanna, J. (2017). Gene expression profiling of patient-derived pancreatic cancer xenografts predicts sensitivity to the BET bromodomain inhibitor JQ1: Implications for individualized medicine efforts. EMBO Molecular Medicine, 9(4), 482–497. https://doi.org/10.15252/emmm.201606975
Zanconato, F., Battilana, G., Forcato, M., Filippi, L., Azzolin, L., Manfrin, A., Quaranta, E., Di Biagio, D., Sigismondo, G., Guzzardo, V., Lejeune, P., Haendler, B., Krijgsveld, J., Fassan, M., Bicciato, S., Cordenonsi, M., & Piccolo, S. (2018). Transcriptional addiction in cancer cells is mediated by YAP/TAZ through BRD4. Nature Medicine, 24(10), 1599–1610. https://doi.org/10.1038/s41591-018-0158-8
Yamazaki, M., Hino, S., Usuki, S., Miyazaki, Y., Oda, T., Nakao, M., Ito, T., & Yamagata, K. (2023). YAP/BRD4-controlled ROR1 promotes tumor-initiating cells and hyperproliferation in pancreatic cancer. EMBO Journal, e112614. https://doi.org/10.15252/embj.2022112614
Tu, M., Klein, L., Espinet, E., Georgomanolis, T., Wegwitz, F., Li, X., Urbach, L., Danieli-Mackay, A., Kuffer, S., Bojarczuk, K., Mizi, A., Gunesdogan, U., Chapuy, B., Gu, Z., Neesse, A., Kishore, U., Strobel, P., Hessmann, E., Hahn, S. A., & Singh, S. K. (2021). TNF-alpha-producing macrophages determine subtype identity and prognosis via AP1 enhancer reprogramming in pancreatic cancer. Nature Cancer, 2(11), 1185–1203. https://doi.org/10.1038/s43018-021-00258-w
Murakami, S., White, S. M., McIntosh, A. T., Nguyen, C. D. K., & Yi, C. (2023). Spontaneously evolved progenitor niches escape Yap oncogene addiction in advanced pancreatic ductal adenocarcinomas. Nature Commununications, 14(1), 1443. https://doi.org/10.1038/s41467-023-37147-y
Scarpa, A., Chang, D. K., Nones, K., Corbo, V., Patch, A. M., Bailey, P., Lawlor, R. T., Johns, A. L., Miller, D. K., Mafficini, A., Rusev, B., Scardoni, M., Antonello, D., Barbi, S., Sikora, K. O., Cingarlini, S., Vicentini, C., McKay, S., Quinn, M. C., & Grimmond, S. M. (2017). Whole-genome landscape of pancreatic neuroendocrine tumours. Nature, 543(7643), 65–71. https://doi.org/10.1038/nature21063
Jiao, Y., Shi, C., Edil, B. H., de Wilde, R. F., Klimstra, D. S., Maitra, A., Schulick, R. D., Tang, L. H., Wolfgang, C. L., Choti, M. A., Velculescu, V. E., Diaz, L. A., Jr., Vogelstein, B., Kinzler, K. W., Hruban, R. H., & Papadopoulos, N. (2011). DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science, 331(6021), 1199–1203. https://doi.org/10.1126/science.1200609
Hackeng, W. M., Brosens, L. A., Poruk, K. E., Noe, M., Hosoda, W., Poling, J. S., Rizzo, A., Campbell-Thompson, M., Atkinson, M. A., Konukiewitz, B., Kloppel, G., Heaphy, C. M., Meeker, A. K., & Wood, L. D. (2016). Aberrant menin expression is an early event in pancreatic neuroendocrine tumorigenesis. Human Pathology, 56, 93–100. https://doi.org/10.1016/j.humpath.2016.06.006
Samdani, R. T., Wasylishen, A. R., Halperin, D. M., Dasari, A., Yao, J. C., Rashid, A., & Estrella, J. S. (2019). Loss of menin expression by immunohistochemistry in pancreatic neuroendocrine tumors: Comparison between primary and metastatic tumors. Pancreas, 48(4), 510–513. https://doi.org/10.1097/MPA.0000000000001274
Xu, J., Ye, Z., Zhuo, Q., Gao, H., Qin, Y., Lou, X., Zhang, W., Wang, F., Wang, Y., Jing, D., Fan, G., Zhang, Y., Chen, X., Chen, J., Xu, X., Yu, X., & Ji, S. (2023). MEN1 degradation induced by neddylation and the CUL4B-DCAF7 axis promotes pancreatic neuroendocrine tumor progression. Cancer Research, 83(13), 2226–2247. https://doi.org/10.1158/0008-5472.CAN-22-3599
Yi, Z., Wei, S., Jin, L., Jeyarajan, S., Yang, J., Gu, Y., Kim, H. S., Schechter, S., Lu, S., Paulsen, M. T., Bedi, K., Narayanan, I. V., Ljungman, M., Crawford, H. C., Pasca di Magliano, M., Ge, K., Dou, Y., & Shi, J. (2022). KDM6A regulates cell plasticity and pancreatic cancer progression by noncanonical activin pathway. Cellular and Molecular Gastroenterology and Hepatology, 13(2), 643–667. https://doi.org/10.1016/j.jcmgh.2021.09.014
Bailey, P., Chang, D. K., Nones, K., Johns, A. L., Patch, A. M., Gingras, M. C., Miller, D. K., Christ, A. N., Bruxner, T. J., Quinn, M. C., Nourse, C., Murtaugh, L. C., Harliwong, I., Idrisoglu, S., Manning, S., Nourbakhsh, E., Wani, S., Fink, L., Holmes, O., & Grimmond, S. M. (2016). Genomic analyses identify molecular subtypes of pancreatic cancer. Nature, 531(7592), 47–52. https://doi.org/10.1038/nature16965
Krauss, L., Urban, B. C., Hastreiter, S., Schneider, C., Wenzel, P., Hassan, Z., Wirth, M., Lankes, K., Terrasi, A., Klement, C., Cernilogar, F. M., Ollinger, R., de Andrade Kratzig, N., Engleitner, T., Schmid, R. M., Steiger, K., Rad, R., Kramer, O. H., Reichert, M., & Schneider, G. (2021). HDAC2 facilitates pancreatic cancer metastasis. Cancer Research. https://doi.org/10.1158/0008-5472.CAN-20-3209
Patil, S., Steuber, B., Kopp, W., Kari, V., Urbach, L., Wang, X., Kuffer, S., Bohnenberger, H., Spyropoulou, D., Zhang, Z., Versemann, L., Bosherz, M. S., Brunner, M., Gaedcke, J., Strobel, P., Zhang, J. S., Neesse, A., Ellenrieder, V., Singh, S. K., & Hessmann, E. (2020). EZH2 regulates pancreatic cancer subtype identity and tumor progression via transcriptional repression of GATA6. Cancer Research, 80(21), 4620–4632. https://doi.org/10.1158/0008-5472.CAN-20-0672
Dandawate, P., Ghosh, C., Palaniyandi, K., Paul, S., Rawal, S., Pradhan, R., Sayed, A. A. A., Choudhury, S., Standing, D., Subramaniam, D., Padhye, S. B., Gunewardena, S., Thomas, S. M., Neil, M. O., Tawfik, O., Welch, D. R., Jensen, R. A., Maliski, S., Weir, S., & Dhar, A. (2019). The histone demethylase KDM3A, increased in human pancreatic tumors, regulates expression of DCLK1 and promotes tumorigenesis in mice. Gastroenterology, 157(6), 1646–1659 e1611. https://doi.org/10.1053/j.gastro.2019.08.018
Kalisz, M., Bernardo, E., Beucher, A., Maestro, M. A., Del Pozo, N., Millan, I., Haeberle, L., Schlensog, M., Safi, S. A., Knoefel, W. T., Grau, V., de Vas, M., Shpargel, K. B., Vaquero, E., Magnuson, T., Ortega, S., Esposito, I., Real, F. X., & Ferrer, J. (2020). HNF1A recruits KDM6A to activate differentiated acinar cell programs that suppress pancreatic cancer. The EMBO Journal, 39(9), e102808. https://doi.org/10.15252/embj.2019102808
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., Sun, Y., Jacobsen, A., Sinha, R., Larsson, E., Cerami, E., Sander, C., & Schultz, N. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling, 6(269), pl1. https://doi.org/10.1126/scisignal.2004088
Cerami, E., Gao, J., Dogrusoz, U., Gross, B. E., Sumer, S. O., Aksoy, B. A., Jacobsen, A., Byrne, C. J., Heuer, M. L., Larsson, E., Antipin, Y., Reva, B., Goldberg, A. P., Sander, C., & Schultz, N. (2012). The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discovery, 2(5), 401–404. https://doi.org/10.1158/2159-8290.CD-12-0095
Barski, A., Cuddapah, S., Cui, K., Roh, T. Y., Schones, D. E., Wang, Z., Wei, G., Chepelev, I., & Zhao, K. (2007). High-resolution profiling of histone methylations in the human genome. Cell, 129(4), 823–837. https://doi.org/10.1016/j.cell.2007.05.009
Bernstein, B. E., Humphrey, E. L., Erlich, R. L., Schneider, R., Bouman, P., Liu, J. S., Kouzarides, T., & Schreiber, S. L. (2002). Methylation of histone H3 Lys 4 in coding regions of active genes. Proceedings of the National Academy of Sciences, 99(13), 8695–8700. https://doi.org/10.1073/pnas.082249499
Bernstein, B. E., Kamal, M., Lindblad-Toh, K., Bekiranov, S., Bailey, D. K., Huebert, D. J., McMahon, S., Karlsson, E. K., Kulbokas, E. J., 3rd, Gingeras, T. R., Schreiber, S. L., & Lander, E. S. (2005). Genomic maps and comparative analysis of histone modifications in human and mouse. Cell, 120(2), 169–181. https://doi.org/10.1016/j.cell.2005.01.001
Heintzman, N. D., Stuart, R. K., Hon, G., Fu, Y., Ching, C. W., Hawkins, R. D., Barrera, L. O., Van Calcar, S., Qu, C., Ching, K. A., Wang, W., Weng, Z., Green, R. D., Crawford, G. E., & Ren, B. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genetics, 39(3), 311–318. https://doi.org/10.1038/ng1966
Sausen, M., Phallen, J., Adleff, V., Jones, S., Leary, R. J., Barrett, M. T., Anagnostou, V., Parpart-Li, S., Murphy, D., Kay Li, Q., Hruban, C. A., Scharpf, R., White, J. R., O’Dwyer, P. J., Allen, P. J., Eshleman, J. R., Thompson, C. B., Klimstra, D. S., Linehan, D. C., & Velculescu, V. E. (2015). Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nature Communications, 6, 7686. https://doi.org/10.1038/ncomms8686
Mozzetta, C., Boyarchuk, E., Pontis, J., & Ait-Si-Ali, S. (2015). Sound of silence: The properties and functions of repressive Lys methyltransferases. Nature Reviews Molecular Cell Biology, 16(8), 499–513. https://doi.org/10.1038/nrm4029
Schulz, W. A., Lang, A., Koch, J., & Greife, A. (2019). The histone demethylase UTX/KDM6A in cancer: Progress and puzzles. International Journal of Cancer, 145(3), 614–620. https://doi.org/10.1002/ijc.32116
Yang, J., Jin, L., Kim, H. S., Tian, F., Yi, Z., Bedi, K., Ljungman, M., Pasca di Magliano, M., Crawford, H., & Shi, J. (2022). KDM6A loss recruits tumor-associated neutrophils and promotes neutrophil extracellular trap formation in pancreatic cancer. Cancer Research, 82(22), 4247–4260. https://doi.org/10.1158/0008-5472.CAN-22-0968
Revia, S., Seretny, A., Wendler, L., Banito, A., Eckert, C., Breuer, K., Mayakonda, A., Lutsik, P., Evert, M., Ribback, S., Gallage, S., Chikh Bakri, I., Breuhahn, K., Schirmacher, P., Heinrich, S., Gaida, M. M., Heikenwalder, M., Calvisi, D. F., Plass, C., & Tschaharganeh, D. F. (2022). Histone H3K27 demethylase KDM6A is an epigenetic gatekeeper of mTORC1 signalling in cancer. Gut, 71(8), 1613–1628. https://doi.org/10.1136/gutjnl-2021-325405
Watanabe, S., Shimada, S., Akiyama, Y., Ishikawa, Y., Ogura, T., Ogawa, K., Ono, H., Mitsunori, Y., Ban, D., Kudo, A., Yamaoka, S., Tanabe, M., & Tanaka, S. (2019). Loss of KDM6A characterizes a poor prognostic subtype of human pancreatic cancer and potentiates HDAC inhibitor lethality. International Journal of Cancer, 145(1), 192–205. https://doi.org/10.1002/ijc.32072
Piunti, A., & Shilatifard, A. (2021). The roles of polycomb repressive complexes in mammalian development and cancer. Nature Reviews. Molecular Cell Biology, 22(5), 326–345. https://doi.org/10.1038/s41580-021-00341-1
Mallen-St Clair, J., Soydaner-Azeloglu, R., Lee, K. E., Taylor, L., Livanos, A., Pylayeva-Gupta, Y., Miller, G., Margueron, R., Reinberg, D., & Bar-Sagi, D. (2012). EZH2 couples pancreatic regeneration to neoplastic progression. Genes & Development, 26(5), 439–444. https://doi.org/10.1101/gad.181800.111
Martinelli, P., Pau, C.-d. S., & E., Cox, T., Sainz, B., Jr., Dusetti, N., Greenhalf, W., Rinaldi, L., Costello, E., Ghaneh, P., Malats, N., Buchler, M., Pajic, M., Biankin, A. V., Iovanna, J., Neoptolemos, J., & Real, F. X. (2017). GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut, 66(9), 1665–1676. https://doi.org/10.1136/gutjnl-2015-311256
Versemann, L., Patil, S., Steuber, B., Zhang, Z., Kopp, W., Krawczyk, H. E., Kaulfuss, S., Wollnik, B., Strobel, P., Neesse, A., Singh, S. K., Ellenrieder, V., & Hessmann, E. (2022). TP53-status-dependent oncogenic EZH2 activity in pancreatic cancer. Cancers (Basel), 14(14). https://doi.org/10.3390/cancers14143451
Chibaya, L., Murphy, K. C., DeMarco, K. D., Gopalan, S., Liu, H., Parikh, C. N., Lopez-Diaz, Y., Faulkner, M., Li, J., Morris, J. P., Ho, Y. J., Chana, S. K., Simon, J., Luan, W., Kulick, A., de Stanchina, E., Simin, K., Zhu, L. J., Fazzio, T. G., & Ruscetti, M. (2023). EZH2 inhibition remodels the inflammatory senescence-associated secretory phenotype to potentiate pancreatic cancer immune surveillance. Nature Cancer. https://doi.org/10.1038/s43018-023-00553-8
Dawkins, J. B., Wang, J., Maniati, E., Heward, J. A., Koniali, L., Kocher, H. M., Martin, S. A., Chelala, C., Balkwill, F. R., Fitzgibbon, J., & Grose, R. P. (2016). Reduced expression of histone methyltransferases KMT2C and KMT2D correlates with improved outcome in pancreatic ductal adenocarcinoma. Cancer Research, 76(16), 4861–4871. https://doi.org/10.1158/0008-5472.CAN-16-0481
Koutsioumpa, M., Hatziapostolou, M., Polytarchou, C., Tolosa, E. J., Almada, L. L., Mahurkar-Joshi, S., Williams, J., Tirado-Rodriguez, A. B., Huerta-Yepez, S., Karavias, D., Kourea, H., Poultsides, G. A., Struhl, K., Dawson, D. W., Donahue, T. R., Fernandez-Zapico, M. E., & Iliopoulos, D. (2019). Lysine methyltransferase 2D regulates pancreatic carcinogenesis through metabolic reprogramming. Gut, 68(7), 1271–1286. https://doi.org/10.1136/gutjnl-2017-315690
Lu, S., Kim, H. S., Cao, Y., Bedi, K., Zhao, L., Narayanan, I. V., Magnuson, B., Gu, Y., Yang, J., Yi, Z., Babaniamansour, S., Shameon, S., Xu, C., Paulsen, M. T., Qiu, P., Jeyarajan, S., Ljungman, M., Thomas, D., Dou, Y., & Shi, J. (2023). KMT2D links TGF-beta signaling to noncanonical activin pathway and regulates pancreatic cancer cell plasticity. International Journal of Cancer. https://doi.org/10.1002/ijc.34528
Li, W., Wu, L., Jia, H., Lin, Z., Zhong, R., Li, Y., Jiang, C., Liu, S., Zhou, X., & Zhang, E. (2021). The low-complexity domains of the KMT2D protein regulate histone monomethylation transcription to facilitate pancreatic cancer progression. Cellular & Molecular Biology Letters, 26(1), 45. https://doi.org/10.1186/s11658-021-00292-7
Yang, G., Yuan, Y., Yuan, H., Wang, J., Yun, H., Geng, Y., Zhao, M., Li, L., Weng, Y., Liu, Z., Feng, J., Bu, Y., Liu, L., Wang, B., & Zhang, X. (2021). Histone acetyltransferase 1 is a succinyltransferase for histones and non-histones and promotes tumorigenesis. EMBO Reports, 22(2), e50967. https://doi.org/10.15252/embr.202050967
Wang, Y., Guo, Y. R., Liu, K., Yin, Z., Liu, R., Xia, Y., Tan, L., Yang, P., Lee, J. H., Li, X. J., Hawke, D., Zheng, Y., Qian, X., Lyu, J., He, J., Xing, D., Tao, Y. J., & Lu, Z. (2017). KAT2A coupled with the alpha-KGDH complex acts as a histone H3 succinyltransferase. Nature, 552(7684), 273–277. https://doi.org/10.1038/nature25003
Liu, J., Shangguan, Y., Tang, D., & Dai, Y. (2021). Histone succinylation and its function on the nucleosome. Journal of Cellular and Molecular Medicine, 25(15), 7101–7109. https://doi.org/10.1111/jcmm.16676
Tong, Y., Guo, D., Yan, D., Ma, C., Shao, F., Wang, Y., Luo, S., Lin, L., Tao, J., Jiang, Y., Lu, Z., & Xing, D. (2020). KAT2A succinyltransferase activity-mediated 14-3-3zeta upregulation promotes beta-catenin stabilization-dependent glycolysis and proliferation of pancreatic carcinoma cells. Cancer Letters, 469, 1–10. https://doi.org/10.1016/j.canlet.2019.09.015
Roth, S. Y., Denu, J. M., & Allis, C. D. (2001). Histone acetyltransferases. Annual Review of Biochemistry, 70, 81–120. https://doi.org/10.1146/annurev.biochem.70.1.81
Ogryzko, V. V., Schiltz, R. L., Russanova, V., Howard, B. H., & Nakatani, Y. (1996). The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87(5), 953–959. https://doi.org/10.1016/s0092-8674(00)82001-2
Garcia-Carpizo, V., Ruiz-Llorente, S., Sarmentero, J., Grana-Castro, O., Pisano, D. G., & Barrero, M. J. (2018). CREBBP/EP300 bromodomains are critical to sustain the GATA1/MYC regulatory axis in proliferation. Epigenetics & Chromatin, 11(1), 30. https://doi.org/10.1186/s13072-018-0197-x
Furlan, T., Kirchmair, A., Sampson, N., Puhr, M., Gruber, M., Trajanoski, Z., Santer, F. R., Parson, W., Handle, F., & Culig, Z. (2021). MYC-mediated ribosomal gene expression sensitizes enzalutamide-resistant prostate cancer cells to EP300/CREBBP inhibitors. The American Journal of Pathology, 191(6), 1094–1107. https://doi.org/10.1016/j.ajpath.2021.02.017
Zhong, Z., Harmston, N., Wood, K. C., Madan, B., & Virshup, D. M. (2022). A p300/GATA6 axis determines differentiation and Wnt dependency in pancreatic cancer models. The Journal of Clinical Investigation, 132(12). https://doi.org/10.1172/JCI156305
Proffitt, K. D., & Virshup, D. M. (2012). Precise regulation of porcupine activity is required for physiological Wnt signaling. The Journal of Biological Chemistry, 287(41), 34167–34178. https://doi.org/10.1074/jbc.M112.381970
Miranda, M., Galli, L. M., Enriquez, M., Szabo, L. A., Gao, X., Hannoush, R. N., & Burrus, L. W. (2014). Identification of the WNT1 residues required for palmitoylation by porcupine. FEBS Letters, 588(24), 4815–4824. https://doi.org/10.1016/j.febslet.2014.11.016
Raphael, B. J., Hruban, R. H., Aguirre, A. J., Moffitt, R. A., Yeh, J. J., Stewart, C., Robertson, A. G., Cherniack, A. D., Gupta, M., Getz, G., & Gabriel, S. B. (2017). Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell, 32(2), 185–203 e113. https://doi.org/10.1016/j.ccell.2017.07.007
Hao, H. X., Xie, Y., Zhang, Y., Charlat, O., Oster, E., Avello, M., Lei, H., Mickanin, C., Liu, D., Ruffner, H., Mao, X., Ma, Q., Zamponi, R., Bouwmeester, T., Finan, P. M., Kirschner, M. W., Porter, J. A., Serluca, F. C., & Cong, F. (2012). ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature, 485(7397), 195–200. https://doi.org/10.1038/nature11019
Koo, B. K., Spit, M., Jordens, I., Low, T. Y., Stange, D. E., van de Wetering, M., van Es, J. H., Mohammed, S., Heck, A. J., Maurice, M. M., & Clevers, H. (2012). Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature, 488(7413), 665–669. https://doi.org/10.1038/nature11308
Jiang, X., Hao, H. X., Growney, J. D., Woolfenden, S., Bottiglio, C., Ng, N., Lu, B., Hsieh, M. H., Bagdasarian, L., Meyer, R., Smith, T. R., Avello, M., Charlat, O., Xie, Y., Porter, J. A., Pan, S., Liu, J., McLaughlin, M. E., & Cong, F. (2013). Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America, 110(31), 12649–12654. https://doi.org/10.1073/pnas.1307218110
Mees, S. T., Mardin, W. A., Wendel, C., Baeumer, N., Willscher, E., Senninger, N., Schleicher, C., Colombo-Benkmann, M., & Haier, J. (2010). EP300--a miRNA-regulated metastasis suppressor gene in ductal adenocarcinomas of the pancreas. International Journal of Cancer, 126(1), 114–124. https://doi.org/10.1002/ijc.24695
Ono, H., Basson, M. D., & Ito, H. (2016). P300 inhibition enhances gemcitabine-induced apoptosis of pancreatic cancer. Oncotarget, 7(32), 51301–51310. https://doi.org/10.18632/oncotarget.10117
Ono, H., Kato, T., Murase, Y., Nakamura, Y., Ishikawa, Y., Watanabe, S., Akahoshi, K., Ogura, T., Ogawa, K., Ban, D., Kudo, A., Akiyama, Y., Tanaka, S., Ito, H., & Tanabe, M. (2021). C646 inhibits G2/M cell cycle-related proteins and potentiates anti-tumor effects in pancreatic cancer. Scientific Reports, 11(1), 10078. https://doi.org/10.1038/s41598-021-89530-8
Arensman, M. D., Telesca, D., Lay, A. R., Kershaw, K. M., Wu, N., Donahue, T. R., & Dawson, D. W. (2014). The CREB-binding protein inhibitor ICG-001 suppresses pancreatic cancer growth. Molecular Cancer Therapeutics, 13(10), 2303–2314. https://doi.org/10.1158/1535-7163.MCT-13-1005
Shi, Y. H., Xu, Q. C., Zhu, Y. Q., Liu, Z. D., Zhao, G. Y., Liu, Q., Wang, X. Y., Wang, J. Q., Xu, X., Su, Q., Lai, J. M., Huang, C. S., & Yin, X. Y. (2023). Imatinib facilitates gemcitabine sensitivity by targeting epigenetically activated PDGFC signaling in pancreatic cancer. Molecular Therapy, 31(2), 503–516. https://doi.org/10.1016/j.ymthe.2022.11.004
Weissmueller, S., Manchado, E., Saborowski, M., Morris, J. P., Wagenblast, E., Davis, C. A., Moon, S. H., Pfister, N. T., Tschaharganeh, D. F., Kitzing, T., Aust, D., Markert, E. K., Wu, J., Grimmond, S. M., Pilarsky, C., Prives, C., Biankin, A. V., & Lowe, S. W. (2014). Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell, 157(2), 382–394. https://doi.org/10.1016/j.cell.2014.01.066
Kurahara, H., Maemura, K., Mataki, Y., Sakoda, M., Shinchi, H., & Natsugoe, S. (2016). Impact of p53 and PDGFR-beta expression on metastasis and prognosis of patients with pancreatic cancer. World Journal of Surgery, 40(8), 1977–1984. https://doi.org/10.1007/s00268-016-3477-2
Principe, D. R., Xiong, R., Li, Y., Pham, T. N. D., Kamath, S. D., Dubrovskyi, O., Ratia, K., Huang, F., Zhao, J., Shen, Z., Thummuri, D., Daohong, Z., Underwood, P. W., Trevino, J., Munshi, H. G., Thatcher, G. R. J., & Rana, A. (2022). XP-524 is a dual-BET/EP300 inhibitor that represses oncogenic KRAS and potentiates immune checkpoint inhibition in pancreatic cancer. Proceedings of the National Academy of Sciences of the United States of America, 119(4). https://doi.org/10.1073/pnas.2116764119
Fan, P., Zhao, J., Meng, Z., Wu, H., Wang, B., Wu, H., & Jin, X. (2019). Overexpressed histone acetyltransferase 1 regulates cancer immunity by increasing programmed death-ligand 1 expression in pancreatic cancer. Journal of Experimental & Clinical Cancer Research, 38(1), 47. https://doi.org/10.1186/s13046-019-1044-z
Chen, C. H., Chuang, S. M., Yang, M. F., Liao, J. W., Yu, S. L., & Chen, J. J. (2012). A novel function of YWHAZ/beta-catenin axis in promoting epithelial-mesenchymal transition and lung cancer metastasis. Molecular Cancer Research, 10(10), 1319–1331. https://doi.org/10.1158/1541-7786.MCR-12-0189
Seto, E., & Yoshida, M. (2014). Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harbor Perspectives in Biology, 6(4), a018713. https://doi.org/10.1101/cshperspect.a018713
Haberland, M., Montgomery, R. L., & Olson, E. N. (2009). The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nature Reviews. Genetics, 10(1), 32–42. https://doi.org/10.1038/nrg2485
Schwer, B., & Verdin, E. (2008). Conserved metabolic regulatory functions of sirtuins. Cell Metabolism, 7(2), 104–112. https://doi.org/10.1016/j.cmet.2007.11.006
Parbin, S., Kar, S., Shilpi, A., Sengupta, D., Deb, M., Rath, S. K., & Patra, S. K. (2014). Histone deacetylases: A saga of perturbed acetylation homeostasis in cancer. The Journal of Histochemistry and Cytochemistry, 62(1), 11–33. https://doi.org/10.1369/0022155413506582
Bannister, A. J., & Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Research, 21(3), 381–395. https://doi.org/10.1038/cr.2011.22
Kelly, R. D., & Cowley, S. M. (2013). The physiological roles of histone deacetylase (HDAC) 1 and 2: Complex co-stars with multiple leading parts. Biochemical Society Transactions, 41(3), 741–749. https://doi.org/10.1042/BST20130010
Shi, X., Hong, T., Walter, K. L., Ewalt, M., Michishita, E., Hung, T., Carney, D., Pena, P., Lan, F., Kaadige, M. R., Lacoste, N., Cayrou, C., Davrazou, F., Saha, A., Cairns, B. R., Ayer, D. E., Kutateladze, T. G., Shi, Y., Cote, J., & Gozani, O. (2006). ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature, 442(7098), 96–99. https://doi.org/10.1038/nature04835
Lehmann, A., Denkert, C., Budczies, J., Buckendahl, A. C., Darb-Esfahani, S., Noske, A., Muller, B. M., Bahra, M., Neuhaus, P., Dietel, M., Kristiansen, G., & Weichert, W. (2009). High class I HDAC activity and expression are associated with RelA/p65 activation in pancreatic cancer in vitro and in vivo. BMC Cancer, 9, 395. https://doi.org/10.1186/1471-2407-9-395
Fritsche, P., Seidler, B., Schuler, S., Schnieke, A., Gottlicher, M., Schmid, R. M., Saur, D., & Schneider, G. (2009). HDAC2 mediates therapeutic resistance of pancreatic cancer cells via the BH3-only protein NOXA. Gut, 58(10), 1399–1409. https://doi.org/10.1136/gut.2009.180711
Cai, M. H., Xu, X. G., Yan, S. L., Sun, Z., Ying, Y., Wang, B. K., & Tu, Y. X. (2018). Depletion of HDAC1, 7 and 8 by histone deacetylase inhibition confers elimination of pancreatic cancer stem cells in combination with gemcitabine. Scientific Reports, 8(1), 1621. https://doi.org/10.1038/s41598-018-20004-0
Shinke, G., Yamada, D., Eguchi, H., Iwagami, Y., Asaoka, T., Noda, T., Wada, H., Kawamoto, K., Gotoh, K., Kobayashi, S., Takeda, Y., Tanemura, M., Mori, M., & Doki, Y. (2018). Role of histone deacetylase 1 in distant metastasis of pancreatic ductal cancer. Cancer Science, 109(8), 2520–2531. https://doi.org/10.1111/cas.13700
Li, J., Wang, J., Wang, J., Nawaz, Z., Liu, J. M., Qin, J., & Wong, J. (2000). Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. The EMBO Journal, 19(16), 4342–4350. https://doi.org/10.1093/emboj/19.16.4342
You, A., Tong, J. K., Grozinger, C. M., & Schreiber, S. L. (2001). CoREST is an integral component of the CoREST- human histone deacetylase complex. Proceedings of the National Academy of Sciences of the United States of America, 98(4), 1454–1458. https://doi.org/10.1073/pnas.98.4.1454
von Burstin, J., Eser, S., Paul, M. C., Seidler, B., Brandl, M., Messer, M., von Werder, A., Schmidt, A., Mages, J., Pagel, P., Schnieke, A., Schmid, R. M., Schneider, G., & Saur, D. (2009). E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology, 137(1), 361–371. https://doi.org/10.1053/j.gastro.2009.04.004
Mishra, V. K., Wegwitz, F., Kosinsky, R. L., Sen, M., Baumgartner, R., Wulff, T., Siveke, J. T., Schildhaus, H. U., Najafova, Z., Kari, V., Kohlhof, H., Hessmann, E., & Johnsen, S. A. (2017). Histone deacetylase class-I inhibition promotes epithelial gene expression in pancreatic cancer cells in a BRD4- and MYC-dependent manner. Nucleic Acids Research, 45(11), 6334–6349. https://doi.org/10.1093/nar/gkx212
Ramaker, R. C., Hardigan, A. A., Gordon, E. R., Wright, C. A., Myers, R. M., & Cooper, S. J. (2021). Pooled CRISPR screening in pancreatic cancer cells implicates co-repressor complexes as a cause of multiple drug resistance via regulation of epithelial-to-mesenchymal transition. BMC Cancer, 21(1), 632. https://doi.org/10.1186/s12885-021-08388-1
Diener, J., Baggiolini, A., Pernebrink, M., Dalcher, D., Lerra, L., Cheng, P. F., Varum, S., Hausel, J., Stierli, S., Treier, M., Studer, L., Basler, K., Levesque, M. P., Dummer, R., Santoro, R., Cantu, C., & Sommer, L. (2021). Epigenetic control of melanoma cell invasiveness by the stem cell factor SALL4. Nature Communications, 12(1), 5056. https://doi.org/10.1038/s41467-021-25326-8
UniProt, C. (2021). UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Research, 49(D1), D480–D489. https://doi.org/10.1093/nar/gkaa1100
Goujon, M., McWilliam, H., Li, W., Valentin, F., Squizzato, S., Paern, J., & Lopez, R. (2010). A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Research, 38(Web Server issue), W695–W699. https://doi.org/10.1093/nar/gkq313
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., & Higgins, D. G. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23(21), 2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Xu, S., Dai, Z., Guo, P., Fu, X., Liu, S., Zhou, L., Tang, W., Feng, T., Chen, M., Zhan, L., Wu, T., Hu, E., Jiang, Y., Bo, X., & Yu, G. (2021). ggtreeExtra: Compact visualization of richly annotated phylogenetic data. Molecular Biology and Evolution, 38(9), 4039–4042. https://doi.org/10.1093/molbev/msab166
Ghandi, M., Huang, F. W., Jane-Valbuena, J., Kryukov, G. V., Lo, C. C., McDonald, E. R., 3rd, Barretina, J., Gelfand, E. T., Bielski, C. M., Li, H., Hu, K., Andreev-Drakhlin, A. Y., Kim, J., Hess, J. M., Haas, B. J., Aguet, F., Weir, B. A., Rothberg, M. V., Paolella, B. R., & Sellers, W. R. (2019). Next-generation characterization of the cancer cell line encyclopedia. Nature, 569(7757), 503–508. https://doi.org/10.1038/s41586-019-1186-3
DepMap, B. (2022). DepMap 22Q4 Public (Version 2). figshare. https://figshare.com/articles/dataset/DepMap_22Q4_Public/21637199/2
Jin, X., Demere, Z., Nair, K., Ali, A., Ferraro, G. B., Natoli, T., Deik, A., Petronio, L., Tang, A. A., Zhu, C., Wang, L., Rosenberg, D., Mangena, V., Roth, J., Chung, K., Jain, R. K., Clish, C. B., Vander Heiden, M. G., & Golub, T. R. (2020). A metastasis map of human cancer cell lines. Nature, 588(7837), 331–336. https://doi.org/10.1038/s41586-020-2969-2
Lechner, S., Malgapo, M. I. P., Gratz, C., Steimbach, R. R., Baron, A., Ruther, P., Nadal, S., Stumpf, C., Loos, C., Ku, X., Prokofeva, P., Lautenbacher, L., Heimburg, T., Wurf, V., Meng, C., Wilhelm, M., Sippl, W., Kleigrewe, K., Pauling, J. K., & Medard, G. (2022). Target deconvolution of HDAC pharmacopoeia reveals MBLAC2 as common off-target. Nature Chemical Biology, 18(8), 812–820. https://doi.org/10.1038/s41589-022-01015-5
Xiong, Y., Donovan, K. A., Eleuteri, N. A., Kirmani, N., Yue, H., Razov, A., Krupnick, N. M., Nowak, R. P., & Fischer, E. S. (2021). Chemo-proteomics exploration of HDAC degradability by small molecule degraders. Cell. Chemistry & Biology, 28(10), 1514–1527 e1514. https://doi.org/10.1016/j.chembiol.2021.07.002
Bond, M. J., Chu, L., Nalawansha, D. A., Li, K., & Crews, C. M. (2020). Targeted degradation of oncogenic KRAS(G12C) by VHL-recruiting PROTACs. ACS Central Science, 6(8), 1367–1375. https://doi.org/10.1021/acscentsci.0c00411
Kaminskas, E., Farrell, A. T., Wang, Y. C., Sridhara, R., & Pazdur, R. (2005). FDA drug approval summary: Azacitidine (5-azacytidine, Vidaza) for injectable suspension. Oncologist, 10(3), 176–182. https://doi.org/10.1634/theoncologist.10-3-176
Kantarjian, H., Issa, J. P., Rosenfeld, C. S., Bennett, J. M., Albitar, M., DiPersio, J., Klimek, V., Slack, J., de Castro, C., Ravandi, F., Helmer, R., 3rd, Shen, L., Nimer, S. D., Leavitt, R., Raza, A., & Saba, H. (2006). Decitabine improves patient outcomes in myelodysplastic syndromes: Results of a phase III randomized study. Cancer, 106(8), 1794–1803. https://doi.org/10.1002/cncr.21792
Barbarotta, L., & Hurley, K. (2015). Romidepsin for the treatment of peripheral T-cell lymphoma. Journal of the Advanced Practitioner in Oncology, 6(1), 22–36 https://www.ncbi.nlm.nih.gov/pubmed/26413372
Mann, B. S., Johnson, J. R., Cohen, M. H., Justice, R., & Pazdur, R. (2007). FDA approval summary: Vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist, 12(10), 1247–1252. https://doi.org/10.1634/theoncologist.12-10-1247
Hoy, S. M. (2020). Tazemetostat: First approval. Drugs, 80(5), 513–521. https://doi.org/10.1007/s40265-020-01288-x
Fraunhoffer, N. A., Moreno Vega, A. I., Abuelafia, A. M., Morvan, M., Lebarbier, E., Mary-Huard, T., Zimmermann, M., Lomberk, G., Urrutia, R., Dusetti, N., Blum, Y., Nicolle, R., & Iovanna, J. (2023). Priming therapy by targeting enhancer-initiated pathways in patient-derived pancreatic cancer cells. EBioMedicine, 92, 104602. https://doi.org/10.1016/j.ebiom.2023.104602
Wong, K. K. (2020). DNMT1 as a therapeutic target in pancreatic cancer: Mechanisms and clinical implications. Cellular Oncology (Dordrecht), 43(5), 779–792. https://doi.org/10.1007/s13402-020-00526-4
Heumann, T. R., Baretti, M., Sugar, E. A., Durham, J. N., Linden, S., Lopez-Vidal, T. Y., Leatherman, J., Cope, L., Sharma, A., Weekes, C. D., O'Dwyer, P. J., Reiss, K. A., Monga, D. K., Ahuja, N., & Azad, N. S. (2022). A randomized, phase II trial of oral azacitidine (CC-486) in patients with resected pancreatic adenocarcinoma at high risk for recurrence. Clinical Epigenetics, 14(1), 166. https://doi.org/10.1186/s13148-022-01367-8
Booth, L., Poklepovic, A., & Dent, P. (2020). Neratinib decreases pro-survival responses of [sorafenib + vorinostat] in pancreatic cancer. Biochemical Pharmacology, 178, 114067. https://doi.org/10.1016/j.bcp.2020.114067
Jo, J. H., Jung, D. E., Lee, H. S., Park, S. B., Chung, M. J., Park, J. Y., Bang, S., Park, S. W., Cho, S., & Song, S. Y. (2022). A phase I/II study of ivaltinostat combined with gemcitabine and erlotinib in patients with untreated locally advanced or metastatic pancreatic adenocarcinoma. International Journal of Cancer, 151(9), 1565–1577. https://doi.org/10.1002/ijc.34144
Fang, Y., McGrail, D. J., Sun, C., Labrie, M., Chen, X., Zhang, D., Ju, Z., Vellano, C. P., Lu, Y., Li, Y., Jeong, K. J., Ding, Z., Liang, J., Wang, S. W., Dai, H., Lee, S., Sahni, N., Mercado-Uribe, I., Kim, T. B., & Mills, G. B. (2019). Sequential therapy with PARP and WEE1 inhibitors minimizes toxicity while maintaining efficacy. Cancer Cell, 35(6), 851–867 e857. https://doi.org/10.1016/j.ccell.2019.05.001
Fernandes Neto, J. M., Nadal, E., Bosdriesz, E., Ooft, S. N., Farre, L., McLean, C., Klarenbeek, S., Jurgens, A., Hagen, H., Wang, L., Felip, E., Martinez-Marti, A., Vidal, A., Voest, E., Wessels, L. F. A., van Tellingen, O., Villanueva, A., & Bernards, R. (2020). Multiple low dose therapy as an effective strategy to treat EGFR inhibitor-resistant NSCLC tumours. Nature Communications, 11(1), 3157. https://doi.org/10.1038/s41467-020-16952-9
Ozkan-Dagliyan, I., Diehl, J. N., George, S. D., Schaefer, A., Papke, B., Klotz-Noack, K., Waters, A. M., Goodwin, C. M., Gautam, P., Pierobon, M., Peng, S., Gilbert, T. S. K., Lin, K. H., Dagliyan, O., Wennerberg, K., Petricoin, E. F., 3rd, Tran, N. L., Bhagwat, S. V., Tiu, R. V., & Der, C. J. (2020). Low-dose vertical inhibition of the RAF-MEK-ERK cascade causes apoptotic death of KRAS mutant cancers. Cell Reports, 31(11), 107764. https://doi.org/10.1016/j.celrep.2020.107764
Wang, H., Cao, Q., & Dudek, A. Z. (2012). Phase II study of panobinostat and bortezomib in patients with pancreatic cancer progressing on gemcitabine-based therapy. Anticancer Research, 32(3), 1027–1031 https://www.ncbi.nlm.nih.gov/pubmed/22399627
Connolly, R. M., Laille, E., Vaishampayan, U., Chung, V., Kelly, K., Dowlati, A., Alese, O. B., Harvey, R. D., Haluska, P., Siu, L. L., Kummar, S., Piekarz, R., Ivy, S. P., Anders, N. M., Downs, M., O’Connor, A., Scardina, A., Saunders, J., Rosner, G. L., & Team, E.-S. (2020). Phase I and pharmacokinetic study of romidepsin in patients with cancer and hepatic dysfunction: A national cancer institute organ dysfunction working group study. Clinical Cancer Research, 26(20), 5329–5337. https://doi.org/10.1158/1078-0432.CCR-20-1412
Chan, E., Arlinghaus, L. R., Cardin, D. B., Goff, L., Berlin, J. D., Parikh, A., Abramson, R. G., Yankeelov, T. E., Hiebert, S., Merchant, N., Bhaskara, S., & Chakravarthy, A. B. (2016). Phase I trial of vorinostat added to chemoradiation with capecitabine in pancreatic cancer. Radiotherapy and Oncology, 119(2), 312–318. https://doi.org/10.1016/j.radonc.2016.04.013
Gounder, M., Schoffski, P., Jones, R. L., Agulnik, M., Cote, G. M., Villalobos, V. M., Attia, S., Chugh, R., Chen, T. W., Jahan, T., Loggers, E. T., Gupta, A., Italiano, A., Demetri, G. D., Ratan, R., Davis, L. E., Mir, O., Dileo, P., Van Tine, B. A., & Stacchiotti, S. (2020). Tazemetostat in advanced epithelioid sarcoma with loss of INI1/SMARCB1: An international, open-label, phase 2 basket study. The Lancet Oncology, 21(11), 1423–1432. https://doi.org/10.1016/S1470-2045(20)30451-4
Sohal, D., Krishnamurthi, S., Tohme, R., Gu, X., Lindner, D., Landowski, T. H., Pink, J., Radivoyevitch, T., Fada, S., Lee, Z., Shepard, D., Khorana, A., & Saunthararajah, Y. (2020). A pilot clinical trial of the cytidine deaminase inhibitor tetrahydrouridine combined with decitabine to target DNMT1 in advanced, chemorefractory pancreatic cancer. American Journal of Cancer Research, 10(9), 3047–3060 https://www.ncbi.nlm.nih.gov/pubmed/33042633
Acknowledgements
Our work was supported by the Deutsche Forschungsgemeinschaft (DFG) (SCHN 959/6-1 and SCHN 959/3-2 to G.S.; SFB1321 (project 329628492) to D.S. and G.S.; KFO5002 to E.H.) and Deutsche Krebshilfe (70113760 to G.S.). Grammar and language were improved by large language models. The authors would like to apologize for any omissions of relevant and important reports in the review article. This may have been due to space limitations, the necessity to selectively choose examples, or inadvertent oversight on our part.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
All authors participated in drafting, conceiving, designing, and writing of the review and revised the manuscript for important intellectual content. All authors approved the final version submitted for publication.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Krauß, L., Schneider, C., Hessmann, E. et al. Epigenetic control of pancreatic cancer metastasis. Cancer Metastasis Rev 42, 1113–1131 (2023). https://doi.org/10.1007/s10555-023-10132-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-023-10132-z