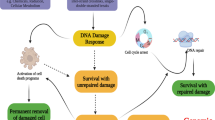
Abstract
The DNA damage, most notably DNA double-strand breaks, poses a serious threat to the stability of mammalian genome. Maintenance of genomic integrity is largely dependent on an efficient, accurate, and timely DNA damage response in the context of chromatin. Consequently, dysregulation of the DNA damage response machinery is fundamentally linked to the genomic instability and a likely predisposition to cancer. In turn, aberrant activation of DNA damage response pathways in human cancers enables tumor cells to survive DNA damages, thus, leading to the development of resistance of tumor cells to DNA damaging radio- and chemotherapies. A substantial body of experimental evidence has established that ATP-dependent chromatin remodeling and histone modifications play a central role in the DNA damage response. As a component of the nucleosome remodeling and histone deacetylase (NuRD) complex that couples both ATP-dependent chromatin remodeling and histone deacetylase activities, the metastasis-associated protein (MTA) family proteins have been recently shown to participate in the DNA damage response beyond its well-established roles in gene transcription. In this thematic review, we will focus on our current understandings of the role of the MTA family proteins in the DNA damage response and their potential implications in DNA damaging anticancer therapy.

Similar content being viewed by others
References
Manavathi, B., & Kumar, R. (2007). Metastasis tumor antigens, an emerging family of multifaceted master coregulators. Journal of Biological Chemistry, 282, 1529–1533.
Manavathi, B., Singh, K., & Kumar, R. (2007). MTA family of coregulators in nuclear receptor biology and pathology. Nuclear Receptor Signaling, 5, e010.
Yang, N., & Xu, R. M. (2013). Structure and function of the BAH domain in chromatin biology. Critical Reviews in Biochemistry and Molecular Biology, 48, 211–221.
Kuo, A. J., Song, J., Cheung, P., Ishibe-Murakami, S., Yamazoe, S., Chen, J. K., et al. (2012). The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature, 484, 115–119.
Onishi, M., Liou, G. G., Buchberger, J. R., Walz, T., & Moazed, D. (2007). Role of the conserved Sir3-BAH domain in nucleosome binding and silent chromatin assembly. Molecular Cell, 28, 1015–1028.
Wang, L., Charroux, B., Kerridge, S., & Tsai, C. C. (2008). Atrophin recruits HDAC1/2 and G9a to modify histone H3K9 and to determine cell fates. EMBO Reports, 9, 555–562.
Ding, Z., Gillespie, L. L., & Paterno, G. D. (2003). Human MI-ER1 alpha and beta function as transcriptional repressors by recruitment of histone deacetylase 1 to their conserved ELM2 domain. Molecular Cell. Biology, 23, 250–258.
Li, S., Paterno, G. D., & Gillespie, L. L. (2013). Nuclear localization of the transcriptional regulator MIER1alpha requires interaction with HDAC1/2 in breast cancer cells. PLoS One, 8, e84046.
Aasland, R., Stewart, A. F., & Gibson, T. (1996). The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends in Biochemical Sciences, 21, 87–88.
Boyer, L. A., Langer, M. R., Crowley, K. A., Tan, S., Denu, J. M., & Peterson, C. L. (2002). Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Molecular Cell, 10, 935–942.
Yu, J., Li, Y., Ishizuka, T., Guenther, M. G., & Lazar, M. A. (2003). A SANT motif in the SMRT corepressor interprets the histone code and promotes histone deacetylation. EMBO Journal, 22, 3403–3410.
Boyer, L. A., Latek, R. R., & Peterson, C. L. (2004). The SANT domain: a unique histone-tail-binding module? Nature Reviews Molecular Cell Biology, 5, 158–163.
Denslow, S. A., & Wade, P. A. (2007). The human Mi-2/NuRD complex and gene regulation. Oncogene, 26, 5433–5438.
Li, D. Q., Pakala, S. B., Nair, S. S., Eswaran, J., & Kumar, R. (2012). Metastasis-associated protein 1/nucleosome remodeling and histone deacetylase complex in cancer. Cancer Research, 72, 387–394.
Lai, A. Y., & Wade, P. A. (2011). Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nature Reviews Cancer, 11, 588–596.
Chou, D. M., Adamson, B., Dephoure, N. E., Tan, X., Nottke, A. C., Hurov, K. E., et al. (2010). A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proceedings of the National Academy of Sciences of the United States of America, 107, 18475–18480.
Li, D. Q., Ohshiro, K., Reddy, S. D., Pakala, S. B., Lee, M. H., Zhang, Y., et al. (2009). E3 ubiquitin ligase COP1 regulates the stability and functions of MTA1. Proceedings of the National Academy of Sciences of the United States of America, 106, 17493–17498.
Li, D. Q., & Kumar, R. (2010). Mi-2/NuRD complex making inroads into DNA-damage response pathway. Cell Cycle, 9, 2071–2079.
Li, D. Q., Divijendra Natha Reddy, S., Pakala, S. B., Wu, X., Zhang, Y., Rayala, S. K., et al. (2009). MTA1 coregulator regulates p53 stability and function. Journal of Biological Chemistry, 284(50), 34545–34552.
Li, D. Q., Ohshiro, K., Khan, M. N., & Kumar, R. (2010). Requirement of MTA1 in ATR-mediated DNA damage checkpoint function. Journal of Biological Chemistry, 285(26), 19802–19812.
Li, D. Q., Pakala, S. B., Reddy, S. D., Ohshiro, K., Peng, S. H., Lian, Y., et al. (2010). Revelation of p53-independent function of MTA1 in DNA damage response via modulation of the p21 WAF1-proliferating cell nuclear antigen pathway. Journal of Biological Chemistry, 285(13), 10044–10052.
Smeenk, G., Wiegant, W. W., Vrolijk, H., Solari, A. P., Pastink, A., & van Attikum, H. (2010). The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. Journal of Cell Biology, 190, 741–749.
Luijsterburg, M. S., & van Attikum, H. (2012). Close encounters of the RNF8th kind: when chromatin meets DNA repair. Current Opinion in Cell Biology, 24, 439–447.
Cann, K. L., & Dellaire, G. (2011). Heterochromatin and the DNA damage response: the need to relax. Biochemical Cell Biology, 89, 45–60.
Toiber, D., Erdel, F., Bouazoune, K., Silberman, D. M., Zhong, L., Mulligan, P., et al. (2013). SIRT6 recruits SNF2H to DNA break sites, preventing genomic instability through chromatin remodeling. Molecular Cell, 51, 454–468.
Jackson, S. P., & Bartek, J. (2009). The DNA-damage response in human biology and disease. Nature, 461(7267), 1071–1078.
Harper, J. W., & Elledge, S. J. (2007). The DNA damage response: ten years after. Molecular Cell, 28, 739–745.
Stucki, M., Clapperton, J. A., Mohammad, D., Yaffe, M. B., Smerdon, S. J., & Jackson, S. P. (2005). MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell, 123, 1213–1226.
Chen, C. C., Carson, J. J., Feser, J., Tamburini, B., Zabaronick, S., Linger, J., et al. (2008). Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell, 134, 231–243.
Hanahan, D., & Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell, 144, 646–674.
Zou, L., & Elledge, S. J. (2003). Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science, 300, 1542–1548.
Vidanes, G. M., Bonilla, C. Y., & Toczyski, D. P. (2005). Complicated tails: histone modifications and the DNA damage response. Cell, 121, 973–976.
Ciccia, A., & Elledge, S. J. (2010). The DNA damage response: making it safe to play with knives. Molecular Cell, 40, 179–204.
Pandita, T. K., & Richardson, C. (2009). Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Research, 37, 1363–1377.
Sulli, G., Di Micco, R., & d’Adda di Fagagna, F. (2012). Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nature Reviews Cancer, 12, 709–720.
Stracker, T. H., & Petrini, J. H. (2011). The MRE11 complex: starting from the ends. Nature Reviews Molecular Cell Biology, 12, 90–103.
Parrilla-Castellar, E. R., Arlander, S. J., & Karnitz, L. (2004). Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair (Amst), 3, 1009–1014.
Lee, J. H., & Paull, T. T. (2004). Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science, 304, 93–96.
Morrison, A. J., Kim, J. A., Person, M. D., Highland, J., Xiao, J., Wehr, T. S., et al. (2007). Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell, 130, 499–511.
Lee, J. H., & Paull, T. T. (2005). ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science, 308, 551–554.
Shiloh, Y., & Ziv, Y. (2013). The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nature Reviews Molecular Cell Biology, 14, 197–210.
Cimprich, K. A., & Cortez, D. (2008). ATR: an essential regulator of genome integrity. Nature Reviews Molecular Cell Biology, 9, 616–627.
Falck, J., Coates, J., & Jackson, S. P. (2005). Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature, 434, 605–611.
Shiloh, Y. (2003). ATM and related protein kinases: safeguarding genome integrity. Nature Reviews Cancer, 3, 155–168.
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S., & Bonner, W. M. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. Journal of Biological Chemistry, 273, 5858–5868.
Stiff, T., O’Driscoll, M., Rief, N., Iwabuchi, K., Lobrich, M., & Jeggo, P. A. (2004). ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Research, 64, 2390–2396.
Ward, I. M., & Chen, J. (2001). Histone H2AX is phosphorylated in an ATR-dependent manner in response to replicational stress. Journal of Biological Chemistry, 276, 47759–47762.
Kruhlak, M. J., Celeste, A., & Nussenzweig, A. (2006). Spatio-temporal dynamics of chromatin containing DNA breaks. Cell Cycle, 5, 1910–1912.
Kinner, A., Wu, W., Staudt, C., & Iliakis, G. (2008). Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Research, 36, 5678–5694.
Soria, G., Polo, S. E., & Almouzni, G. (2012). Prime, repair, restore: the active role of chromatin in the DNA damage response. Molecular Cell, 46, 722–734.
Paull, T. T., Rogakou, E. P., Yamazaki, V., Kirchgessner, C. U., Gellert, M., & Bonner, W. M. (2000). A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Current Biology, 10, 886–895.
Morrison, A. J., Highland, J., Krogan, N. J., Arbel-Eden, A., Greenblatt, J. F., Haber, J. E., et al. (2004). INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell, 119, 767–775.
van Attikum, H., Fritsch, O., Hohn, B., & Gasser, S. M. (2004). Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell, 119, 777–788.
Zhang, R., Liu, S. T., Chen, W., Bonner, M., Pehrson, J., Yen, T. J., et al. (2007). HP1 proteins are essential for a dynamic nuclear response that rescues the function of perturbed heterochromatin in primary human cells. Molecular Cell. Biology, 27, 949–962.
Murga, M., Jaco, I., Fan, Y., Soria, R., Martinez-Pastor, B., Cuadrado, M., et al. (2007). Global chromatin compaction limits the strength of the DNA damage response. Journal of Cell Biology, 178, 1101–1108.
Xu, Y., Sun, Y., Jiang, X., Ayrapetov, M. K., Moskwa, P., Yang, S., et al. (2010). The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. Journal of Cell Biology, 191, 31–43.
Takahashi, K., & Kaneko, I. (1985). Changes in nuclease sensitivity of mammalian cells after irradiation with 60Co gamma-rays. International Journal of Radiation Biology and Related Studies in Physics, Chemistry, and Medicine, 48, 389–395.
Ziv, Y., Bielopolski, D., Galanty, Y., Lukas, C., Taya, Y., Schultz, D. C., et al. (2006). Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nature Cell Biology, 8, 870–876.
Price, B. D., & D’Andrea, A. D. (2013). Chromatin remodeling at DNA double-strand breaks. Cell, 152, 1344–1354.
Altaf, M., Saksouk, N., & Cote, J. (2007). Histone modifications in response to DNA damage. Mutation Research, 618, 81–90.
Bao, Y. (2011). Chromatin response to DNA double-strand break damage. Epigenomics, 3, 307–321.
Huertas, D., Sendra, R., & Munoz, P. (2009). Chromatin dynamics coupled to DNA repair. Epigenetics, 4, 31–42.
Lai, W., Li, H., Liu, S., & Tao, Y. (2013). Connecting chromatin modifying factors to DNA damage response. International Journal of Molecular Sciences, 14, 2355–2369.
Lans, H., Marteijn, J. A., & Vermeulen, W. (2012). ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics & Chromatin, 5, 4.
Luijsterburg, M. S., & van Attikum, H. (2011). Chromatin and the DNA damage response: the cancer connection. Molecular Oncology, 5, 349–367.
Mendez-Acuna, L., Di Tomaso, M. V., Palitti, F., & Martinez-Lopez, W. (2010). Histone post-translational modifications in DNA damage response. Cytogenetic and Genome Research, 128, 28–36.
Smeenk, G., & van Attikum, H. (2013). The chromatin response to DNA breaks: leaving a mark on genome integrity. Annual Review of Biochemistry, 82, 55–80.
Park, J. H., Park, E. J., Lee, H. S., Kim, S. J., Hur, S. K., Imbalzano, A. N., et al. (2006). Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO Journal, 25, 3986–3997.
Clapier, C. R., & Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annual Review of Biochemistry, 78, 273–304.
Vignali, M., Hassan, A. H., Neely, K. E., & Workman, J. L. (2000). ATP-dependent chromatin-remodeling complexes. Molecular Cell. Biology, 20, 1899–1910.
Bao, Y., & Shen, X. (2007). SnapShot: chromatin remodeling complexes. Cell, 129, 632.
Papamichos-Chronakis, M., Krebs, J. E., & Peterson, C. L. (2006). Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes and Development, 20, 2437–2449.
Liang, B., Qiu, J., Ratnakumar, K., & Laurent, B. C. (2007). RSC functions as an early double-strand-break sensor in the cell’s response to DNA damage. Current Biology, 17, 1432–1437.
Seeber, A., Dion, V., & Gasser, S. M. (2013). Checkpoint kinases and the INO80 nucleosome remodeling complex enhance global chromatin mobility in response to DNA damage. Genes and Development, 27, 1999–2008.
Klochendler-Yeivin, A., Picarsky, E., & Yaniv, M. (2006). Increased DNA damage sensitivity and apoptosis in cells lacking the Snf5/Ini1 subunit of the SWI/SNF chromatin remodeling complex. Molecular Cell. Biology, 26, 2661–2674.
Park, J. H., Park, E. J., Hur, S. K., Kim, S., & Kwon, J. (2009). Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA Repair (Amst), 8, 29–39.
van Attikum, H., Fritsch, O., & Gasser, S. M. (2007). Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO Journal, 26, 4113–4125.
Chai, B., Huang, J., Cairns, B. R., & Laurent, B. C. (2005). Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes and Development, 19, 1656–1661.
Das, C., Lucia, M. S., Hansen, K. C., & Tyler, J. K. (2009). CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature, 459, 113–117.
Masumoto, H., Hawke, D., Kobayashi, R., & Verreault, A. (2005). A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature, 436, 294–298.
Bhaskara, S., Knutson, S. K., Jiang, G., Chandrasekharan, M. B., Wilson, A. J., Zheng, S., et al. (2010). Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell, 18, 436–447.
Dovey, O. M., Foster, C. T., Conte, N., Edwards, S. A., Edwards, J. M., Singh, R., et al. (2013). Histone deacetylase 1 and 2 are essential for normal T-cell development and genomic stability in mice. Blood, 121, 1335–1344.
Miller, K. M., Tjeertes, J. V., Coates, J., Legube, G., Polo, S. E., Britton, S., et al. (2010). Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nature Structural and Molecular Biology, 17, 1144–1151.
Wang, R. H., Sengupta, K., Li, C., Kim, H. S., Cao, L., Xiao, C., et al. (2008). Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell, 14, 312–323.
Lee, H. S., Park, J. H., Kim, S. J., Kwon, S. J., & Kwon, J. (2010). A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO Journal, 29, 1434–1445.
Xue, Y., Wong, J., Moreno, G. T., Young, M. K., Cote, J., & Wang, W. (1998). NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Molecular Cell, 2, 851–861.
Zhang, Y., LeRoy, G., Seelig, H. P., Lane, W. S., & Reinberg, D. (1998). The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell, 95, 279–289.
Tong, J. K., Hassig, C. A., Schnitzler, G. R., Kingston, R. E., & Schreiber, S. L. (1998). Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature, 395, 917–921.
Fujita, N., Jaye, D. L., Kajita, M., Geigerman, C., Moreno, C. S., & Wade, P. A. (2003). MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell, 113, 207–219.
Polo, S. E., Kaidi, A., Baskcomb, L., Galanty, Y., & Jackson, S. P. (2010). Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. [Research Support, Non-U.S. Gov’t]. EMBO Journal, 29(18), 3130–3139. doi:10.1038/emboj.2010.188.
Smeenk, G., & van Attikum, H. (2011). NuRD alert! NuRD regulates the DNA damage response. Epigenomics, 3, 133–135.
Larsen, D. H., Poinsignon, C., Gudjonsson, T., Dinant, C., Payne, M. R., Hari, F. J., et al. (2010). The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. Journal of Cell Biology, 190, 731–740.
Schmidt, D. R., & Schreiber, S. L. (1999). Molecular association between ATR and two components of the nucleosome remodeling and deacetylating complex, HDAC2 and CHD4. Biochemistry, 38, 14711–14717.
Dornan, D., Shimizu, H., Mah, A., Dudhela, T., Eby, M., O’Rourke, K., et al. (2006). ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science, 313, 1122–1126.
Tanaka, H., Arakawa, H., Yamaguchi, T., Shiraishi, K., Fukuda, S., Matsui, K., et al. (2000). A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature, 404, 42–49.
Ward, I. M., Minn, K., & Chen, J. (2004). UV-induced ataxia-telangiectasia-mutated and Rad3-related (ATR) activation requires replication stress. Journal of Biological Chemistry, 279, 9677–9680.
Liu, Q., Guntuku, S., Cui, X. S., Matsuoka, S., Cortez, D., Tamai, K., et al. (2000). Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes and Development, 14, 1448–1459.
Guo, Z., Kumagai, A., Wang, S. X., & Dunphy, W. G. (2000). Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes and Development, 14, 2745–2756.
Chini, C. C., & Chen, J. (2003). Human claspin is required for replication checkpoint control. Journal of Biological Chemistry, 278, 30057–30062.
van Haaften, G., Romeijn, R., Pothof, J., Koole, W., Mullenders, L. H., Pastink, A., et al. (2006). Identification of conserved pathways of DNA-damage response and radiation protection by genome-wide RNAi. Current Biology, 16, 1344–1350.
Errico, A., Aze, A., & Costanzo, V. (2014). Mta2 promotes Tipin-dependent maintenance of replication fork integrity. Cell Cycle, 13, 2120–2128.
Ayrapetov, M. K., Xu, C., Sun, Y., Zhu, K., Parmar, K., D’Andrea, A. D., et al. (2011). Activation of Hif1alpha by the prolylhydroxylase inhibitor dimethyoxalyglycine decreases radiosensitivity. PLoS One, 6, e26064.
Thurn, K. T., Thomas, S., Raha, P., Qureshi, I., & Munster, P. N. (2013). Histone deacetylase regulation of ATM-mediated DNA damage signaling. Molecular Cancer Therapeutics, 12, 2078–2087.
Bhaskara, S., Jacques, V., Rusche, J. R., Olson, E. N., Cairns, B. R., & Chandrasekharan, M. B. (2013). Histone deacetylases 1 and 2 maintain S-phase chromatin and DNA replication fork progression. Epigenetics & Chromatin, 6, 27.
O’Shaughnessy, A., & Hendrich, B. (2013). CHD4 in the DNA-damage response and cell cycle progression: not so NuRDy now. Biochemical Society Transactions, 41, 777–782.
Pan, M. R., Hsieh, H. J., Dai, H., Hung, W. C., Li, K., Peng, G., et al. (2012). Chromodomain helicase DNA-binding protein 4 (CHD4) regulates homologous recombination DNA repair, and its deficiency sensitizes cells to poly(ADP-ribose) polymerase (PARP) inhibitor treatment. Journal of Biological Chemistry, 287, 6764–6772.
Luijsterburg, M. S., Acs, K., Ackermann, L., Wiegant, W. W., Bekker-Jensen, S., Larsen, D. H., et al. (2012). A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO Journal, 31, 2511–2527.
Sims, J. K., & Wade, P. A. (2011). Mi-2/NuRD complex function is required for normal S phase progression and assembly of pericentric heterochromatin. Molecular Biology of the Cell, 22, 3094–3102.
Zhao, S., Choi, M., Overton, J. D., Bellone, S., Roque, D. M., Cocco, E., et al. (2013). Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proceedings of the National Academy of Sciences of the United States of America, 110, 2916–2921.
Wu, M., Wang, L., Li, Q., Li, J., Qin, J., & Wong, J. (2013). The MTA family proteins as novel histone H3 binding proteins. Cell Bioscience, 3, 1.
Goodarzi, A. A., Noon, A. T., Deckbar, D., Ziv, Y., Shiloh, Y., Lobrich, M., et al. (2008). ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Molecular Cell, 31, 167–177.
Kumar, R., Wang, R. A., & Bagheri-Yarmand, R. (2003). Emerging roles of MTA family members in human cancers. Seminar in Oncology, 30, 30–37.
Yu, L., Su, Y. S., Zhao, J., Wang, H., & Li, W. (2013). Repression of NR4A1 by a chromatin modifier promotes docetaxel resistance in PC-3 human prostate cancer cells. FEBS Letters, 587, 2542–2551.
Acknowledgments
We are in debt to our colleagues in this field whose original work may have not been cited here due to space limitations. This study was supported by the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (No. 2013–06) (to D-Q L) and NIH grants CA98823 (to RK).
Conflict of interest
The authors declare no any potential conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, DQ., Yang, Y. & Kumar, R. MTA family of proteins in DNA damage response: mechanistic insights and potential applications. Cancer Metastasis Rev 33, 993–1000 (2014). https://doi.org/10.1007/s10555-014-9524-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-014-9524-2