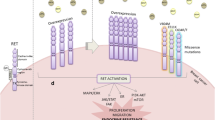
Abstract
Purpose
Resistance to endocrine therapy is the primary cause of treatment failure and death in patients with ER-positive (ER +)/luminal breast cancer. Expression and activation of the RET receptor tyrosine kinase may be driving poor outcomes. We aim to identify high-risk patients and druggable pathways for biomarker-based clinical trials.
Methods
We obtained batch-normalized mRNA expression data from Breast Invasive Carcinoma-The Cancer Genome Atlas, PanCancer Atlas (BRCA-TCGA). To determine clinically significant cutoffs for RET expression, patients were grouped at different thresholds for Kaplan–Meier plotting. Differential gene expression (DGE) analysis and enrichment for gene sets was performed. transcriptomic dataset of antiestrogen-treated ER + tumors stratified by clinical response was then analyzed.
Results
High RET expression was associated with worse outcomes in patients with ER + tumors, and stratification was enhanced by incorporating GDNF expression. High RET/GDNF patients had significantly lower overall survival (HR = 2.04, p = 0.012), progression-free survival (HR = 2.87, p < 0.001), disease-free survival (HR = 2.67, p < 0.001), and disease-specific survival (HR = 3.53, p < 0.001) than all other ER + patients. High RET/GDNF tumors were enriched for estrogen-independent signaling and targetable pathways including NTRK, PI3K, and KRAS. Tumors with adaptive resistance to endocrine therapy were enriched for gene expression signatures of high RET/GDNF primary tumors.
Conclusion
Expression and activation of the RET receptor tyrosine kinase may be driving poor outcomes in some patients with ER + breast cancer. ER + patients above the 75th percentile may benefit from clinical trials with tyrosine kinase inhibitors.







Similar content being viewed by others
Data availability
The datasets used and code generated during the current study are available on a public GitHub repository (www.github.com/rashatk/HighRETGDNF). The dataset obtained from Xia et al. can be accessed at https://doi.org/10.1158/1078-0432.CCR-21-3189.
References
Rondón-Lagos M, Villegas V, Rangel N, Sánchez M, Zaphiropoulos P (2016) Tamoxifen resistance: emerging molecular targets. IJMS 17(8):1357
Lei S, Zheng R, Zhang S, Wang S, Chen R, Sun K, Zeng H, Zhou J, Wei W (2021) Global patterns of breast cancer incidence and mortality: a population-based cancer registry data analysis from 2000 to 2020. Cancer Commun (Lond) 41(11):1183–1194
Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale AL (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98(19):10869–10874
Hanker AB, Sudhan DR, Arteaga CL (2020) Overcoming endocrine resistance in breast cancer. Cancer Cell 37(4):496–513
Liu X-r, Zhang R-y, Gong H, Rugo HS, Chen L-b, Yuan F, Che J-w, Tie J, Shao B, Wan F-l, Kong W-y, Song G-h, Jiang H-f, Guo-bing X, Li H-p (2020) Methylome variation predicts exemestane resistance in advanced ER + breast cancer. Technol Cancer Res Treat 19:153303381989633
Razavi P, Chang MT, Xu G, Bandlamudi C, Ross DS, Vasan N, Cai Y, Bielski CM, Donoghue MTA, Jonsson P, Penson A, Shen R, Pareja F, Kundra R, Middha S, Cheng ML, Zehir A, Kandoth C, Patel R, Huberman K, Smyth LM, Jhaveri K, Modi S, Traina TA, Dang C, Zhang W, Weigelt B, Li BT, Ladanyi M, Hyman DM, Schultz N, Robson ME, Hudis C, Brogi E, Viale A, Norton L, Dickler MN, Berger MF, Iacobuzio-Donahue CA, Chandarlapaty S, Scaltriti M, Reis-Filho JS, Solit DB, Taylor BS, Baselga J (2018) The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell 34(3):427-438.e6
Xia Y, He X, Renshaw L, Martinez-Perez C, Kay C, Gray M, Meehan J, Parker JS, Perou CM, Carey LA, Dixon JM, Turnbull A (2022) Integrated DNA and RNA sequencing reveals drivers of endocrine resistance in estrogen receptor-positive breast cancer. Clin Cancer Res 28(16):3618–3629
Plaza-Menacho I, Morandi A, Robertson D, Pancholi S, Drury S, Dowsett M, Martin LA, Isacke CM (2010) Targeting the receptor tyrosine kinase RET sensitizes breast cancer cells to tamoxifen treatment and reveals a role for RET in endocrine resistance. Oncogene 29(33):4648–4657
Spanheimer PM, Cyr AR, Gillum MP, Woodfield GW, Askeland RW, Weigel RJ (2014) Distinct pathways regulated by RET and estrogen receptor in luminal breast cancer demonstrate the biological basis for combination therapy. Ann Surg 259(4):793–799
Spanheimer PM, Park JM, Askeland RW, Kulak MV, Woodfield GW, De Andrade JP, Cyr AR, Sugg SL, Thomas A, Weigel RJ (2014) Inhibition of ret increases the efficacy of antiestrogen and is a novel treatment strategy for luminal breast cancer. Clin Cancer Res 20(8):2115–2125
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2(5):401–404
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci Signal [Internet]. 2013 Apr 2 [cited 2022 Oct 22];6 (269). Available from: https://www.science.org/doi/https://doi.org/10.1126/scisignal.2004088
Zhao X, Rødland EA, Tibshirani R, Plevritis S (2015) Molecular subtyping for clinically defined breast cancer subgroups. Breast Cancer Res 17(1):29
GDNF Protein Expression Summary [Internet]. The Human Protein Atlas. Available from: https://www.proteinatlas.org/ENSG00000168621-GDNF
GDNF [Internet]. NCBI Gene Testing Registry. Available from: https://www.ncbi.nlm.nih.gov/gtr/genes/2668/
Therneau T. A Package for Survival Analysis in R [Internet] [Internet]. 2020. Available from: https://CRAN.R-project.org/package=survival
survminer: Survival Analysis and Visualization [Internet]. Available from: https://rpkgs.datanovia.com/survminer/index.html
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550
Gu Z, Eils R, Schlesner M (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32(18):2847–2849
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, Feng T, Zhou L, Tang W, Zhan L, Fu X, Liu S, Bo X, Yu G (2021) clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation 2(3):100141
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102(43):15545–15550
Wickham H (2016) ggplot2: elegant graphics for data analysis, 2nd edn. Springer International Publishing, Cham
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504
Tarazona S, Furió-Tarí P, Turrà D, Pietro AD, Nueda MJ, Ferrer A, Conesa A (2015) Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res 16:gkv711
Foroutan M, Bhuva DD, Lyu R, Horan K, Cursons J, Davis MJ (2018) Single sample scoring of molecular phenotypes. BMC Bioinform 19(1):404
Clayton NS, Grose RP (2018) Emerging roles of fibroblast growth factor 10 in cancer. Front Genet 24(9):499
Zou Y, Xie J, Tian W, Wu L, Xie Y, Huang S, Tang Y, Deng X, Wu H, Xie X (2022) Integrative analysis of KCNK genes and establishment of a specific prognostic signature for breast cancer. Front Cell Dev Biol 17(10):839986
Alvarez-Baron CP, Jonsson P, Thomas C, Dryer SE, Williams C (2011) The two-pore domain potassium channel KCNK5: induction by estrogen receptor α and role in proliferation of breast cancer cells. Mol Endocrinol 25(8):1326–1336
Valla M, Klæstad E, Ytterhus B, Bofin AM (2022) CCND1 Amplification in breast cancer -associations with proliferation, histopathological grade, molecular subtype and prognosis. J Mammary Gland Biol Neoplasia 27(1):67–77
Astashchanka A, Shroka TM, Jacobsen BM (2019) Mucin 2 (MUC2) modulates the aggressiveness of breast cancer. Breast Cancer Res Treat 173(2):289–299
Wang X, Wang S (2021) Identification of key genes involved in tamoxifen-resistant breast cancer using bioinformatics analysis. Transl Cancer Res TCR 10(12):5246–5257
Lakshmanan I, Ponnusamy MP, Das S, Chakraborty S, Haridas D, Mukhopadhyay P, Lele SM, Batra SK (2012) MUC16 induced rapid G2/M transition via interactions with JAK2 for increased proliferation and anti-apoptosis in breast cancer cells. Oncogene 31(7):805–817
Liu J, Wang X, Sun J, Chen Y, Li J, Huang J, Du H, Gan L, Qiu Z, Li H, Ren G, Wei Y (2022) The novel methylation biomarker npy5r sensitizes breast cancer cells to chemotherapy. Front Cell Dev Biol 11(9):798221
Zheng X, Zhang Y, Qi X, Wang M, Sun P, Zhang Y, Jiang J (2014) Role of Sphk1 in the malignant transformation of breast epithelial cells and breast cancer progression. Indian J Cancer 51(4):524
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC (2003) PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34(3):267–273
Wang J, Xu Y, Li L, Wang L, Yao R, Sun Q, Du G (2017) FOXC1 is associated with estrogen receptor alpha and affects sensitivity of tamoxifen treatment in breast cancer. Cancer Med 6(1):275–287
Gaudet MM, Chanock S, Dunning A, Driver K, Brinton LA, Lissowska J, Peplonska B, Pharoah P, Garcia-Closas M (2008) HSD17B1 genetic variants and hormone receptor-defined breast cancer. Cancer Epidemiol Biomark Prev 17(10):2766–2772
Jin TY, Saindane M, Park KS, Kim S, Nam S, Yoo Y, Yang JH, Yun I (2021) LEP as a potential biomarker in prognosis of breast cancer: systemic review and meta analyses (PRISMA). Medicine 100(33):e26896
Liu J, Welm B, Boucher KM, Ebbert MTW, Bernard PS (2012) TRIM29 functions as a tumor suppressor in nontumorigenic breast cells and invasive ER+ breast cancer. Am J Pathol 180(2):839–847
Yin L, Li Q, Mrdenovic S, Chu GCY, Wu BJ, Bu H, Duan P, Kim J, You S, Lewis MS, Liang G, Wang R, Zhau HE, Chung LWK (2022) KRT13 promotes stemness and drives metastasis in breast cancer through a plakoglobin/c-Myc signaling pathway. Breast Cancer Res 24(1):7
National Library of Medicine Gene Database [Internet]. Available from: https://www.ncbi.nlm.nih.gov/gene/?term=
Guo X, Ramirez I, Garcia YA, Velasquez EF, Gholkar AA, Cohn W, Whitelegge JP, Tofig B, Damoiseaux R, Torres JZ (2021) DUSP7 regulates the activity of ERK2 to promote proper chromosome alignment during cell division. J Biol Chem 296:100676
Li L, Wang N, Xiong Y, Guo G, Zhu M, Gu Y (2022) Transcription factor FOSL1 enhances drug resistance of breast cancer through DUSP7-mediated dephosphorylation of PEA15. Mol Cancer Res 20(4):515–526
Adhikari H, Kattan WE, Kumar S, Zhou P, Hancock JF, Counter CM (2021) Oncogenic KRAS is dependent upon an EFR3A-PI4KA signaling axis for potent tumorigenic activity. Nat Commun 12(1):5248
Liu H, Wen T, Zhou Y, Fan X, Du T, Gao T, Li L, Liu J, Yang L, Yao J, Ge Y, An G (2019) DCLK1 plays a metastatic-promoting role in human breast cancer cells. Biomed Res Int 15(2019):1–8
Sundvall M, Iljin K, Kilpinen S, Sara H, Kallioniemi OP, Elenius K (2008) Role of ErbB4 in breast cancer. J Mammary Gland Biol Neoplasia 13(2):259–268
Chao MW, Lin TE, HuangFu WC, Chang CD, Tu HJ, Chen LC, Yen SC, Sung TY, Huang WJ, Yang CR, Pan SL, Hsu KC (2021) Identification of a dual TAOK1 and MAP4K5 inhibitor using a structure-based virtual screening approach. J Enzyme Inhib Med Chem 36(1):98–108
Lo HW, Hsu SC, Hung MC (2006) EGFR signaling pathway in breast cancers: from traditional signal transduction to direct nuclear translocalization. Breast Cancer Res Treat 95(3):211–218
Bose D, Banerjee S, Singh RK, Wise LM, Robertson ES (2020) Vascular endothelial growth factor encoded by Parapoxviruses can regulate metabolism and survival of triple negative breast cancer cells. Cell Death Dis 11(11):996
Gao C, Li H, Zhuang J, Zhang H, Wang K, Yang J, Liu C, Liu L, Zhou C, Sun C (2018) The construction and analysis of ceRNA networks in invasive breast cancer: a study based on The Cancer Genome Atlas. CMAR 11:1–11
Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, Nathenson M, Doebele RC, Farago AF, Pappo AS, Turpin B, Dowlati A, Brose MS, Mascarenhas L, Federman N, Berlin J, El-Deiry WS, Baik C, Deeken J, Boni V, Nagasubramanian R, Taylor M, Rudzinski ER, Meric-Bernstam F, Sohal DPS, Ma PC, Raez LE, Hechtman JF, Benayed R, Ladanyi M, Tuch BB, Ebata K, Cruickshank S, Ku NC, Cox MC, Hawkins DS, Hong DS, Hyman DM (2018) Efficacy of Larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med 378(8):731–739
Joshi SK, Qian K, Bisson WH, Watanabe-Smith K, Huang A, Bottomly D, Traer E, Tyner JW, McWeeney SK, Davare MA, Druker BJ, Tognon CE (2020) Discovery and characterization of targetable NTRK point mutations in hematologic neoplasms. Blood 135(24):2159–2170
Bera A, Subramanian M, Karaian J, Eklund M, Radhakrishnan S, Gana N, Rothwell S, Pollard H, Hu H, Shriver CD, Srivastava M (2020) Functional role of vitronectin in breast cancer. Ramos JW, editor. PLoS ONE 15(11):0242141
Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL (2001) Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J Biol Chem 276(17):13615–13621
Bourdeau V, Deschênes J, Métivier R, Nagai Y, Nguyen D, Bretschneider N, Gannon F, White JH, Mader S (2004) Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol 18(6):1411–1427
Hallett RM, Hassell JA (2014) Estrogen independent gene expression defines clinically relevant subgroups of estrogen receptor positive breast cancer. BMC Cancer 14(1):871
Spanheimer PM, Woodfield GW, Cyr AR, Kulak MV, White-Baer LS, Bair TB, Weigel RJ (2013) Expression of the RET Proto-oncogene is regulated by TFAP2C in breast cancer independent of the estrogen receptor. Ann Surg Oncol 20(7):2204–2212
Miller KD, Trigo JM, Wheeler C, Barge A, Rowbottom J, Sledge G, Baselga J (2005) A multicenter phase II trial of ZD6474, a vascular endothelial growth factor receptor-2 and epidermal growth factor receptor tyrosine kinase inhibitor, in patients with previously treated metastatic breast cancer. Clin Cancer Res 11(9):3369–3376
Yardley DA, Dees EC, Myers SD, Li S, Healey P, Wang Z, Brickman MJ, Paolini J, Kern KA, Citrin DL (2012) Phase II open-label study of sunitinib in patients with advanced breast cancer. Breast Cancer Res Treat 136(3):759–767
Mayer EL, Isakoff SJ, Klement G, Downing SR, Chen WY, Hannagan K, Gelman R, Winer EP, Burstein HJ (2012) Combination antiangiogenic therapy in advanced breast cancer: a phase 1 trial of vandetanib, a VEGFR inhibitor, and metronomic chemotherapy, with correlative platelet proteomics. Breast Cancer Res Treat 136(1):169–178
Boér K, Láng I, Llombart-Cussac A, Andreasson I, Vivanco GL, Sanders N, Pover GM, Murray E (2012) Vandetanib with docetaxel as second-line treatment for advanced breast cancer: a double-blind, placebo-controlled, randomized Phase II study. Invest New Drugs 30(2):681–687
Matias-Guiu X, Calabuig R, Badia F, Serra J, La Calle JP (1988) Spontaneous infarcts in fibroadenomas of the breast. Curr Surg 45(4):277–279
Lim JSJ, Wong ALA, Ow SGW, Ngoi NYL, Chan GHJ, Ang YLE, Chong WQ, Lim SE, Lim YW, Lee M, Choo JRE, Tan HL, Yong WP, Soo RA, Tan DSP, Chee CE, Sundar R, Yadav K, Jain S, Wang L, Tai BC, Goh BC, Lee SC (2022) Phase Ib/II dose expansion study of lenvatinib combined with letrozole in postmenopausal women with hormone receptor-positive breast cancer. Clin Cancer Res 28(11):2248–2256
W-xin P, J-guo H, Yang L, A-hua G, Mo YY (2017) Linc-RoR promotes MAPK/ERK signaling and confers estrogen-independent growth of breast cancer. Mol Cancer 16(1):161
Ferrando L, Vingiani A, Garuti A, Vernieri C, Belfiore A, Agnelli L, Dagrada G, Ivanoiu D, Bonizzi G, Munzone E, Lippolis L, Dameri M, Ravera F, Colleoni M, Viale G, Magnani L, Ballestrero A, Zoppoli G, Pruneri G (2023) ESR1 gene amplification and MAP3K mutations are selected during adjuvant endocrine therapies in relapsing Hormone Receptor-positive, HER2-negative breast cancer (HR+ HER2- BC). PLoS Genet 19(1):e1010563
Spanheimer PM, Bashir A, Lorenzen AW, Beck AC, Liao J, Lizarraga IM, Erdahl LM, Sugg SL, Karwal MW, Weigel RJ (2021) A pilot study of preoperative vandetanib on markers of proliferation and apoptosis in breast cancer. Am J Clin Oncol 44(9):456–462
Acknowledgements
This work was presented, in part, as an oral presentation at the 18th Annual Academic Surgical Congress, February 8, 2023 in Houston, TX.
Funding
Dr Philip Spanheimer is supported by the National Institutes of Health grant P50CA058223. This work was supported by a UNC Lineberger Comprehensive Cancer Center Developmental Award which is supported in part by P30 CA016086 Cancer Center Core Support Grant.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by RTK. Project visualization and planning was performed by RTK and PMS. Writing of the original draft and editing the manuscript was performed by RTK, HK, AW, and PMS. Project supervision and funding management was performed by PMS. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no potential conflicts of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kakati, R.T., Kim, H., Whitman, A. et al. High expression of the RET receptor tyrosine kinase and its ligand GDNF identifies a high-risk subset of estrogen receptor positive breast cancer. Breast Cancer Res Treat 199, 589–601 (2023). https://doi.org/10.1007/s10549-023-06937-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-023-06937-9