Abstract
Purpose
Hormone receptor-positive (HR+) and human epidermal growth factor receptor 2 negative (HER2−) early breast cancer (BC) is the most prevalent BC subtype with substantial biological heterogeneity. Although clinicopathological (CP) characteristics have a clear prognostic value, additional biomarkers could refine survival prediction and guide treatment decision.
Methods
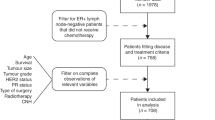
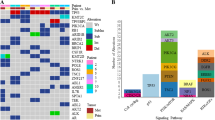
Copy number aberrations and somatic driver mutations were obtained with OncoScan CGH array and sequencing of 36 genes on HR+/HER2− node-positive early BC patients treated with chemotherapy from the PACS04 trial. We built a two-gene genomic score (GS) associated with distant disease-free survival (DDFS), whose prognostic value was assessed on the external METABRIC data (n = 1413) using overall survival (OS) and breast cancer-specific survival (BCSS).
Results
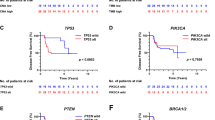
In the PACS04 trial (n = 327), the median follow-up for DDFS (65 events) was 9.6 years. FGFR1 amplifications (\({\text{HR}}_{\text{Amplification}}\) = 2.44, 95% CI [1.25; 4.76], p = 0.009) and MAP3K1 mutations (\({\text{HR}}_{\text{Mutation}}\) = 0.10, [0.01; 0.78], p = 0.03) were associated with DDFS beyond CP characteristics. A prognostic GS combining FGFR1 amplifications and MAP3K1 mutations added more information to CP model (\(\chi_{\text{DDFS}}^{2}\) = 12.97, \(p_{\text{DDFS}}\) < 0.001 and \(\chi_{\text{OS}}^{2}\) = 11.52, \(p_{\text{OS}}\) < 0.001). In the METABRIC study (n = 1413), FGFR1 amplifications (\({\text{HR}}_{\text{Amplification}}\) = 2.00 [1.40; 2.87], p < 0.001) and MAP3K1 mutations (\({\text{HR}}_{\text{Mutation}}\) = 0.58 [0.41; 0.83], p = 0.003) were significantly associated with BCSS beyond CP characteristics. The prognostic GS added significant prognostic information to CP model (\(\chi_{\text{BCSSS}}^{2}\) = 15.39, \(p_{\text{BCSS}}\) < 0.001 and \(\chi_{\text{OS}}^{2}\) = 5.62, \(p_{\text{OS}}\) = 0.02).
Conclusion
In axillary node-positive, HR+, and HER2− early BC, amplifications of FGFR1 gene were strongly associated with increased risk for distant disease, while mutations of MAP3K1 gene were significantly associated with decreased risk.








Similar content being viewed by others
References
Howlader N, Altekruse SF, Li CI, Chen VW, Clarke CA, Ries LAG et al (2014) US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J Natl Cancer Inst. https://doi.org/10.1093/jnci/dju055
Cianfrocca M, Goldstein LJ (2004) Prognostic and predictive factors in early-stage breast cancer. Oncologist 9:606–616
Rugo HS (2008) The importance of distant metastases in hormone-sensitive breast cancer. Breast 17(Suppl 1):S3–S8
Rosano GMC (2007) The importance of distant metastasis and its impact on survival rates on early-stage hormone receptor-positive breast cancer. US Oncol Dis 1:22–25
Mohammadizadeh F, Hani M, Ranaee M, Bagheri M (2013) Role of cyclin D1 in breast carcinoma. J Res Med Sci 18:1021–1025
Roy PG, Pratt N, Purdie CA, Baker L, Ashfield A, Quinlan P et al (2010) High CCND1 amplification identifies a group of poor prognosis women with estrogen receptor positive breast cancer. Int J Cancer 127:355–360
Turner N, Pearson A, Sharpe R, Lambros M, Geyer F, Lopez-Garcia MA et al (2010) FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res 70:2085–2094
Drago JZ, Niemierko A, Spring L, Moy B, Juric D, Isakoff SJ et al (2017) FGFR gene amplification and response to endocrine therapy in metastatic hormone receptor positive (HR+) breast cancer. JCO 35:1013
Vendrell JA, Thollet A, Nguyen NT, Ghayad SE, Vinot S, Bièche I et al (2012) ZNF217 is a marker of poor prognosis in breast cancer that drives epithelial-mesenchymal transition and invasion. Cancer Res 72:3593–3606
Littlepage LE, Adler AS, Kouros-Mehr H, Huang G, Chou J, Krig SR et al (2012) The transcription factor ZNF217 is a prognostic biomarker and therapeutic target during breast cancer progression. Cancer Discov 2:638–651
Krig SR, Miller JK, Frietze S, Beckett LA, Neve RM, Farnham PJ et al (2010) ZNF217, a candidate breast cancer oncogene amplified at 20q13, regulates expression of the ErbB3 receptor tyrosine kinase in breast cancer cells. Oncogene 29:5500–5510
Kim Y-S, Won YS, Park KS, Song BJ, Kim JS, Oh SJ et al (2008) Prognostic significance of HER2 gene amplification according to stage of breast cancer. J Korean Med Sci 23:414–420
Borg A, Baldetorp B, Fernö M, Killander D, Olsson H, Sigurdsson H (1991) ERBB2 amplification in breast cancer with a high rate of proliferation. Oncogene 6:137–143
Carr JA, Havstad S, Zarbo RJ, Divine G, Mackowiak P, Velanovich V (2000) The association of HER-2/neu amplification with breast cancer recurrence. Arch Surg 135:1469–1474
Letessier A, Sircoulomb F, Ginestier C, Cervera N, Monville F, Gelsi-Boyer V et al (2006) Frequency, prognostic impact, and subtype association of 8p12, 8q24, 11q13, 12p13, 17q12, and 20q13 amplifications in breast cancers. BMC Cancer 6:245
Cizkova M, Susini A, Vacher S, Cizeron-Clairac G, Andrieu C, Driouch K et al (2012) PIK3CA mutation impact on survival in breast cancer patients and in ERα, PR and ERBB2-based subgroups. Breast Cancer Res 14:R28
Fernández-Cuesta L, Oakman C, Falagan-Lotsch P, Smoth K-S, Quinaux E, Buyse M et al (2012) Prognostic and predictive value of TP53 mutations in node-positive breast cancer patients treated with anthracycline- or anthracycline/taxane-based adjuvant therapy: results from the BIG 02-98 phase III trial. Breast Cancer Res 14:R70
Pham TT, Angus SP, Johnson GL (2013) MAP3K1: genomic alterations in cancer and function in promoting cell survival or apoptosis. Genes Cancer 4:419–426
Network Cancer Genome Atlas (2012) Comprehensive molecular portraits of human breast tumours. Nature 490:61–70
Roché H, Allouache D, Romieu G, Bourgeois H, Canon J, Serin D et al (2009) Five-year analysis of the FNCLCC-PACS04 trial: FEC100 vs ED75 for the adjuvant treatment of node positive breast cancer. Cancer Res 69:602
Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ et al (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486:346–352
Commo F, Guinney J, Ferté C, Bot B, Lefebvre C, Soria J-C et al (2016) rCGH: a comprehensive array-based genomic profile platform for precision medicine. Bioinformatics 32:1402–1404
Adélaïde J, Finetti P, Bekhouche I, Repellini L, Geneix J, Sircoulomb F et al (2007) Integrated profiling of basal and luminal breast cancers. Cancer Res 67:11565–11575
Blanche P. TimeROC: time-dependent ROC curve and AUC for censored survival data. https://cran.r-project.org/web/packages/timeROC/timeROC.pdf
Gelsi-Boyer V, Orsetti B, Cervera N, Finetti P, Sircoulomb F, Rougé C et al (2005) Comprehensive profiling of 8p11-12 amplification in breast cancer. Mol Cancer Res 3:655–667
Griffith OL, Spies NC, Anurag M, Griffith M, Luo J, Tu D et al (2018) The prognostic effects of somatic mutations in ER-positive breast cancer. Nat Commun 9:3476
Acknowledgements
We thank Yuki Takahashi for some editorial assistance.
Funding
This study was funded by grants from ANR and CGI (RHU MyPROBE and ANR-17-RHUS-0008) and Inca (PAIR SEIN14-049 IDF-ANDRE).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no relevant conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the PACS04 study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Carene, D., Tran-Dien, A., Lemonnier, J. et al. Association between FGFR1 copy numbers, MAP3K1 mutations, and survival in axillary node-positive, hormone receptor-positive, and HER2-negative early breast cancer in the PACS04 and METABRIC studies. Breast Cancer Res Treat 179, 387–401 (2020). https://doi.org/10.1007/s10549-019-05462-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-019-05462-y