Abstract
Purpose
This study aimed to elucidate the clinicopathological characteristics of breast tumors with homologous recombination deficiency (HRD) and the sensitivity to neoadjuvant paclitaxel followed by fluorouracil, epirubicin, and cyclophosphamide (P-FEC).
Methods
Tumor biopsy samples obtained before P-FEC from 141 patients with stages II–III breast cancer including the luminal (n = 76), luminal-HER2 (n = 13), HER2 (n = 17), and triple-negative (TNBC, n = 35) subtypes were subjected to assay for HRD score using the OncoScan CNV FFPE Assay Kit. HRD score was a simple sum of NtAI, LOH, and LST (cutoff, 42). TNBCs were also subjected to the gene expression assay using the Affymetrix microarray (U133 plus 2.0) and to the BRCA1 promoter methylation assay using the methylation-specific real-time PCR.
Results
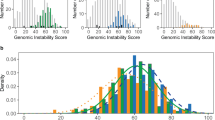
Of the 141 breast tumors, 45 samples (32%) had high HRD scores and were associated with high histological grade (P = 0.001), negative progesterone receptor (P = 0.018), high Ki67 index (P = 0.032), and BRCA1 promoter methylation (P = 3.6e−07). The proportion of tumors with high HRD scores was significantly higher in the TNBC subtype than the others (P = 0.006). In the TNBC subtype, but not the others, high HRD scores were significantly (P = 0.001) associated with a low pathological complete response rate to P-FEC. Among the molecular TNBC subtypes, a majority of tumors belonging to the basal-like 1, immunomodulatory, mesenchymal, mesenchymal stem-like, but not luminal androgen receptor (LAR), subtypes had high HRD scores.
Conclusions
Approximately one-third of sporadic breast tumors show a high HRD score, indicating the presence of homologous recombination dysfunction, and they are characterized by biologically aggressive phenotypes, most commonly in the TNBC subtype, and less sensitive to P-FEC.



Similar content being viewed by others
Abbreviations
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- HER2:
-
Human epidermal growth factor receptor 2
- TNBC:
-
Triple-negative breast cancer
- TILs:
-
Tumor-infiltrating lymphocytes
- HG:
-
Histological grade
- IHC:
-
Immunohistochemistry
- pCR:
-
Pathologic complete response
References
Krejci L, Altmannova V, Spirek M, Zhao X (2012) Homologous recombination and its regulation. Nucleic Acids Res 40(13):5795–5818. https://doi.org/10.1093/nar/gks270
Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, Delaloge S, Li W, Tung N, Armstrong A, Wu W, Goessl C, Runswick S, Conte P (2017) Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 377(6):523–533. https://doi.org/10.1056/NEJMoa1706450
Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC, Szallasi Z, Barry WT, Winer EP, Tung NM, Isakoff SJ, Ryan PD, Greene-Colozzi A, Gutin A, Sangale Z, Iliev D, Neff C, Abkevich V, Jones JT, Lanchbury JS, Hartman AR, Garber JE, Ford JM, Silver DP, Richardson AL (2016) Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res 22(15):3764–3773. https://doi.org/10.1158/1078-0432.CCR-15-2477
Birkbak NJ, Wang ZC, Kim JY, Eklund AC, Li Q, Tian R, Bowman-Colin C, Li Y, Greene-Colozzi A, Iglehart JD, Tung N, Ryan PD, Garber JE, Silver DP, Szallasi Z, Richardson AL (2012) Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov 2(4):366–375. https://doi.org/10.1158/2159-8290.CD-11-0206
Popova T, Manié E, Rieunier G, Caux-Moncoutier V, Tirapo C, Dubois T, Delattre O, Sigal-Zafrani B, Bollet M, Longy M, Houdayer C, Sastre-Garau X, Vincent-Salomon A, Stoppa-Lyonnet D, Stern MH (2012) Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res 72(21):5454–5462. https://doi.org/10.1158/0008-5472.CAN-12-1470
Abkevich V, Timms KM, Hennessy BT, Potter J, Carey MS, Meyer LA, Smith-McCune K, Broaddus R, Lu KH, Chen J, Tran TV, Williams D, Iliev D, Jammulapati S, FitzGerald LM, Krivak T, DeLoia JA, Gutin A, Mills GB, Lanchbury JS (2012) Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer 107(10):1776–1782. https://doi.org/10.1038/bjc.2012.451
Kaklamani VG, Jeruss JS, Hughes E, Siziopikou K, Timms KM, Gutin A, Abkevich V, Sangale Z, Solimeno C, Brown KL, Jones J, Hartman AR, Meservey C, Jovanovic B, Helenowski I, Khan SA, Bethke K, Hansen N, Uthe R, Giordano S, Rosen S, Hoskins K, Von Roenn J, Jain S, Parini V, Gradishar W (2015) Phase II neoadjuvant clinical trial of carboplatin and eribulin in women with triple negative early-stage breast cancer (NCT01372579). Breast Cancer Res Treat 151(3):629–638. https://doi.org/10.1007/s10549-015-3435-y
Loibl S, Weber KE, Timms KM, Elkin EP, Hahnen E, Fasching PA, Lederer B, Denkert C, Schneeweiss A, Braun S, Salat CT, Rezai M, Blohmer JU, Zahm DM, Jackisch C, Gerber B, Klare P, Kümmel S, Schem C, Paepke S, Schmutzler R, Rhiem K, Penn S, Reid J, Nekljudova V, Hartman AR, von Minckwitz G, Untch M (2018) Survival analysis of carboplatin added to an anthracycline/taxane-based neoadjuvant chemotherapy and HRD score as predictor of response—final results from GeparSixto. Ann. Oncol. https://doi.org/10.1093/annonc/mdy460
Telli ML, Metzger O, Timms K, Evans B, Vogel D, Wei H, Jones JT, Wenstrup RJ, McKee MD, Sullivan DM (2018) Evaluation of homologous recombination deficiency (HRD) status with pathological response to carboplatin+/−veliparib in BrighTNess, a randomized phase 3 study in early stage TNBC. J Clin Oncol. 36(15_suppl):519. https://doi.org/10.1200/JCO.2018.36.15_suppl.519
Tutt A, Tovey H, Cheang MCU, Kernaghan S, Kilburn L, Gazinska P, Owen J, Abraham J, Barrett S, Barrett-Lee P, Brown R, Chan S, Dowsett M, Flanagan JM, Fox L, Grigoriadis A, Gutin A, Harper-Wynne C, Hatton MQ, Hoadley KA, Parikh J, Parker P, Perou CM, Roylance R, Shah V, Shaw A, Smith IE, Timms KM, Wardley AM, Wilson G, Gillett C, Lanchbury JS, Ashworth A, Rahman N, Harries M, Ellis P, Pinder SE, Bliss JM (2018) Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med 24(5):628–637. https://doi.org/10.1038/s41591-018-0009-7
Hahnen E, Lederer B, Hauke J, Loibl S, Kröber S, Schneeweiss A, Denkert C, Fasching PA, Blohmer JU, Jackisch C, Paepke S, Gerber B, Kümmel S, Schem C, Neidhardt G, Huober J, Rhiem K, Costa S, Altmüller J, Hanusch C, Thiele H, Müller V, Nürnberg P, Karn T, Nekljudova V, Untch M, von Minckwitz G, Schmutzler RK (2017) Germline mutation status, pathological complete response, and disease-free survival in triple-negative breast cancer: secondary analysis of the GeparSixto randomized clinical trial. JAMA Oncol 3(10):1378–1385. https://doi.org/10.1001/jamaoncol.2017.1007
Wang C, Zhang J, Wang Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, Xie Y (2015) Prevalence of BRCA1 mutations and responses to neoadjuvant chemotherapy among BRCA1 carriers and non-carriers with triple-negative breast cancer. Ann Oncol 26(3):523–528. https://doi.org/10.1093/annonc/mdu559
Pfeifer W, Sokolenko AP, Potapova ON, Bessonov AA, Ivantsov AO, Laptiev SA, Zaitseva OA, Yatsuk OS, Matsko DE, Semiglazova TY, Togo AV, Imyanitov EN (2014) Breast cancer sensitivity to neoadjuvant therapy in BRCA1 and CHEK2 mutation carriers and non-carriers. Breast Cancer Res Treat 148(3):675–683. https://doi.org/10.1007/s10549-014-3206-1
Byrski T, Huzarski T, Dent R, Marczyk E, Jasiowka M, Gronwald J, Jakubowicz J, Cybulski C, Wisniowski R, Godlewski D, Lubinski J, Narod SA (2014) Pathologic complete response to neoadjuvant cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res Treat 147(2):401–405. https://doi.org/10.1007/s10549-014-3100-x
Byrski T, Gronwald J, Huzarski T, Grzybowska E, Budryk M, Stawicka M, Mierzwa T, Szwiec M, Wisniowski R, Siolek M, Dent R, Lubinski J, Narod S (2010) Pathologic complete response rates in young women with BRCA1-positive breast cancers after neoadjuvant chemotherapy. J Clin Oncol 28(3):375–379. https://doi.org/10.1200/JCO.2008.20.7019
Byrski T, Huzarski T, Dent R, Gronwald J, Zuziak D, Cybulski C, Kladny J, Gorski B, Lubinski J, Narod SA (2009) Response to neoadjuvant therapy with cisplatin in BRCA1-positive breast cancer patients. Breast Cancer Res Treat 115(2):359–363. https://doi.org/10.1007/s10549-008-0128-9
Loibl S, O’Shaughnessy J, Untch M, Sikov WM, Rugo HS, McKee MD, Huober J, Golshan M, von Minckwitz G, Maag D, Sullivan D, Wolmark N, McIntyre K, Ponce Lorenzo JJ, Metzger Filho O, Rastogi P, Symmans WF, Liu X, Geyer CE (2018) Addition of the PARP inhibitor veliparib plus carboplatin or carboplatin alone to standard neoadjuvant chemotherapy in triple-negative breast cancer (BrighTNess): a randomised, phase 3 trial. Lancet Oncol 19(4):497–509. https://doi.org/10.1016/S1470-2045(18)30111-6
Akashi-Tanaka S, Watanabe C, Takamaru T, Kuwayama T, Ikeda M, Ohyama H, Mori M, Yoshida R, Hashimoto R, Terumasa S, Enokido K, Hirota Y, Okuyama H, Nakamura S (2015) BRCAness predicts resistance to taxane-containing regimens in triple negative breast cancer during neoadjuvant chemotherapy. Clin Breast Cancer 15(1):80–85. https://doi.org/10.1016/j.clbc.2014.08.003
Tanino H, Kosaka Y, Nishimiya H, Tanaka Y, Minatani N, Kikuchi M, Shida A, Waraya M, Katoh H, Enomoto T, Sengoku N, Kajita S, Hoffman RM, Watanabe M (2016) BRCAness and prognosis in triple-negative breast cancer patients treated with neoadjuvant chemotherapy. PLoS ONE 11(12):e0165721. https://doi.org/10.1371/journal.pone.0165721
Lips EH, Mulder L, Hannemann J, Laddach N, Vrancken Peeters MT, van de Vijver MJ, Wesseling J, Nederlof PM, Rodenhuis S (2011) Indicators of homologous recombination deficiency in breast cancer and association with response to neoadjuvant chemotherapy. Ann Oncol 22(4):870–876. https://doi.org/10.1093/annonc/mdq468
Telli ML, Hellyer J, Audeh W, Jensen KC, Bose S, Timms KM, Gutin A, Abkevich V, Peterson RN, Neff C, Hughes E, Sangale Z, Jones J, Hartman AR, Chang PJ, Vinayak S, Wenstrup R, Ford JM (2018) Homologous recombination deficiency (HRD) status predicts response to standard neoadjuvant chemotherapy in patients with triple-negative or BRCA1/2 mutation-associated breast cancer. Breast Cancer Res Treat 168(3):625–630. https://doi.org/10.1007/s10549-017-4624-7
Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, Ramakrishna M, Martin S, Boyault S, Sieuwerts AM, Simpson PT, King TA, Raine K, Eyfjord JE, Kong G, Borg Å, Birney E, Stunnenberg HG, van de Vijver MJ, Børresen-Dale AL, Martens JW, Span PN, Lakhani SR, Vincent-Salomon A, Sotiriou C, Tutt A, Thompson AM, Van Laere S, Richardson AL, Viari A, Campbell PJ, Stratton MR, Nik-Zainal S (2017) HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 23(4):517–525. https://doi.org/10.1038/nm.4292
Marquard AM, Eklund AC, Joshi T, Krzystanek M, Favero F, Wang ZC, Richardson AL, Silver DP, Szallasi Z, Birkbak NJ (2015) Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark Res 3:9. https://doi.org/10.1186/s40364-015-0033-4
Timms KM, Abkevich V, Hughes E, Neff C, Reid J, Morris B, Kalva S, Potter J, Tran TV, Chen J, Iliev D, Sangale Z, Tikishvili E, Perry M, Zharkikh A, Gutin A, Lanchbury JS (2014) Association of BRCA1/2 defects with genomic scores predictive of DNA damage repair deficiency among breast cancer subtypes. Breast Cancer Res 16(6):475. https://doi.org/10.1186/s13058-014-0475-x
Naoi Y, Kishi K, Tanei T, Tsunashima R, Tominaga N, Baba Y, Kim SJ, Taguchi T, Tamaki Y, Noguchi S (2011) Prediction of pathologic complete response to sequential paclitaxel and 5-fluorouracil/epirubicin/cyclophosphamide therapy using a 70-gene classifier for breast cancers. Cancer 117(16):3682–3690. https://doi.org/10.1002/cncr.25953
Bareche Y, Venet D, Ignatiadis M, Aftimos P, Piccart M, Rothe F, Sotiriou C (2018) Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann Oncol 29(4):895–902. https://doi.org/10.1093/annonc/mdy024
Van Loo P, Nordgard SH, Lingjærde OC, Russnes HG, Rye IH, Sun W, Weigman VJ, Marynen P, Zetterberg A, Naume B, Perou CM, Børresen-Dale AL, Kristensen VN (2010) Allele-specific copy number analysis of tumors. Proc Natl Acad Sci USA 107(39):16910–16915. https://doi.org/10.1073/pnas.1009843107
Morimoto K, Kim SJ, Tanei T, Shimazu K, Tanji Y, Taguchi T, Tamaki Y, Terada N, Noguchi S (2009) Stem cell marker aldehyde dehydrogenase 1-positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression. Cancer Sci 100(6):1062–1068. https://doi.org/10.1111/j.1349-7006.2009.01151.x
Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, Wienert S, Van den Eynden G, Baehner FL, Penault-Llorca F, Perez EA, Thompson EA, Symmans WF, Richardson AL, Brock J, Criscitiello C, Bailey H, Ignatiadis M, Floris G, Sparano J, Kos Z, Nielsen T, Rimm DL, Allison KH, Reis-Filho JS, Loibl S, Sotiriou C, Viale G, Badve S, Adams S, Willard-Gallo K, Loi S, 2014 ITWG (2015) The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol 26(2):259–271. https://doi.org/10.1093/annonc/mdu450
Iwamoto T, Yamamoto N, Taguchi T, Tamaki Y, Noguchi S (2011) BRCA1 promoter methylation in peripheral blood cells is associated with increased risk of breast cancer with BRCA1 promoter methylation. Breast Cancer Res Treat 129(1):69–77. https://doi.org/10.1007/s10549-010-1188-1
Winter C, Nilsson MP, Olsson E, George AM, Chen Y, Kvist A, Törngren T, Vallon-Christersson J, Hegardt C, Häkkinen J, Jönsson G, Grabau D, Malmberg M, Kristoffersson U, Rehn M, Gruvberger-Saal SK, Larsson C, Borg Å, Loman N, Saal LH (2016) Targeted sequencing of BRCA1 and BRCA2 across a large unselected breast cancer cohort suggests that one-third of mutations are somatic. Ann Oncol 27(8):1532–1538. https://doi.org/10.1093/annonc/mdw209
Wei M, Grushko TA, Dignam J, Hagos F, Nanda R, Sveen L, Xu J, Fackenthal J, Tretiakova M, Das S, Olopade OI (2005) BRCA1 promoter methylation in sporadic breast cancer is associated with reduced BRCA1 copy number and chromosome 17 aneusomy. Cancer Res 65(23):10692–10699. https://doi.org/10.1158/0008-5472.CAN-05-1277
Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, Wilfond B, Borg A, Trent J, Raffeld M, Yakhini Z, Ben-Dor A, Dougherty E, Kononen J, Bubendorf L, Fehrle W, Pittaluga S, Gruvberger S, Loman N, Johannsson O, Olsson H, Sauter G (2001) Gene-expression profiles in hereditary breast cancer. N Engl J Med 344(8):539–548. https://doi.org/10.1056/NEJM200102223440801
Zhang L, Long X (2015) Association of BRCA1 promoter methylation with sporadic breast cancers: Evidence from 40 studies. Sci Rep 5:17869. https://doi.org/10.1038/srep17869
Foulkes WD (2006) BRCA1 and BRCA2: chemosensitivity, treatment outcomes and prognosis. Fam Cancer 5(2):135–142. https://doi.org/10.1007/s10689-005-2832-5
Isakoff SJ, Mayer EL, He L, Traina TA, Carey LA, Krag KJ, Rugo HS, Liu MC, Stearns V, Come SE, Timms KM, Hartman AR, Borger DR, Finkelstein DM, Garber JE, Ryan PD, Winer EP, Goss PE, Ellisen LW (2015) TBCRC009: a multicenter phase II clinical trial of platinum monotherapy with biomarker assessment in metastatic triple-negative breast cancer. J Clin Oncol 33(17):1902–1909. https://doi.org/10.1200/JCO.2014.57.6660
Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA (2011) Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121(7):2750–2767. https://doi.org/10.1172/JCI45014
Acknowledgements
The authors would like to express special thanks to Dr. Nicolai Juul Birkbak for valuable advice on the determination of HRD scores. This study was supported in part by AstraZeneca Externally Sponsored Research [Tracking Number NCR-16-12580] and Affymetrix Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Shinzaburo Noguchi has been an adviser for Taiho, AstraZeneca, and Novartis and has received research funding for this study from AstraZeneca and for other studies from Sysmex, Novartis, Chugai, Daiichi-Sankyo, Kyowa-Kirin, Takeda, Pfizer, Ono, Taiho, and Eisai and honoraria from AstraZeneca, Novartis, Pfizer, Chugai, Takeda, Sysmex, Nippon Kayaku, and Ono. Yasuto Naoi has received research funding for this study from AstraZeneca and honoraria from Sysmex. Naofumi Kagara has received honoraria from AstraZeneca and Novartis. Masafumi Shimoda has received research funding for other studies from Novartis and AstraZeneca and honoraria from Chugai, Eisai, Novartis, and Takeda. Kenzo Shimazu has received honoraria from AstraZeneca, Chugai, and Sysmex. Seung Jim Kim has received honoraria from AstraZeneca, Chugai, Eisai, Kyowa-Kirin, Novartis, Pfizer, Shimadzu, Taiho, and Takeda. The other authors declare no conflicts of interest.
Ethical approval
This study complies with the current relevant laws of and guidelines for Japan.
Informed consent
The study protocol was approved by the Ethical Review Board of Osaka University Hospital, and informed consent was obtained from each patient before tumor biopsy.
Rights and permissions
About this article
Cite this article
Imanishi, S., Naoi, Y., Shimazu, K. et al. Clinicopathological analysis of homologous recombination-deficient breast cancers with special reference to response to neoadjuvant paclitaxel followed by FEC. Breast Cancer Res Treat 174, 627–637 (2019). https://doi.org/10.1007/s10549-018-05120-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-018-05120-9