Abstract
Reconstructing ancestral species is a challenging endeavour: fossils are often scarce or enigmatic, and inferring ancestral characters based on novel molecular approaches (e.g. comparative genomics or developmental genetics) has long been controversial. A key philosophical challenge pertinent at present is the lack of a theoretical framework capable of evaluating inferences of homology made through integration of multiple kinds of evidence (e.g. molecular, developmental, or morphological). Here, I present just such a framework. I start with a brief history and critical assessment of attempts at inferring morphological homology through developmental genetics. I then bring attention to a recent model of homology, namely Character Identity Mechanisms (DiFrisco et al. 2020), intended partly to elucidate the relationships between morphological characters, developmental genetics, and homology. I utilise and build on this model to construct the evaluative framework mentioned above, which judges the epistemic value of evidence of each kind in each particular case based on three proposed criteria: effectiveness, admissibility, and informativity, as well as providing a generalised guideline on how it can be scientifically operationalised. I then point out the evolution of the eumetazoan body plan as a case in point where the application of this framework can yield satisfactory results, both empirically and conceptually. I will conclude with a discussion on some potential implications for more general philosophy of biology and philosophy of science, especially surrounding evidential integration, models and explanation, and reductionism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction: finding homology in the deeps
Reconstructing characters and character states of extinct ancestral organisms faces the general challenge of every historical science: making inferences about things that are no longer directly observable. There are few research areas where this challenge manifests itself as strongly as in the reconstruction of early animals: this area of research—crucial to understanding how metazoan complexity has evolved—suffers from scarcity of fossil evidence, and where such evidence exists it tends to be enigmatic and hard to place within a broader context, largely because candidate metazoan Precambrian fossils are not easily comparable to most living animals (Budd 2015). Furthermore, comparative morphology is in dispute over key early metazoan characters, especially the general body plan and its more specialised derivations (Genikhovich and Technau 2017; Nielsen 2019; Nielsen et al. 2018). Another way to phrase this challenge is to describe it as discerning homologous from non-homologous characters in the absence of extensive evidence. Is segmentation an ancestral bilaterian character (homology), or did it evolve independently in different taxa (non-homology)? Is the bilaterian antero-posterior axis homologous to the anthozoan directive axis or the oral-aboral axis, or neither? Is larval morphology conserved across metazoan evolution or are different larvae the result of convergent evolution?
The introduction of molecular approaches to discerning homology and all the evidence associated with them has not comprehensively solved the problem either. This is unsurprising, as introducing new evidence and methodology often adds as many problems as it solves; these new problems share with the original ones only the explanandum. It has long been recognised that phenomena studied directly by developmental genetics, such as patterns of gene expression or cis- and trans-regulatory processes, are not always strongly associated across phylogeny with the morphological characters they are deemed to causally underlie (Abouheif 1997; Haag 2014; Nielsen and Martinez 2003; True and Haag 2001; Wagner 2014; Wray 2007; Wray and Abouheif 1998). This has led some authors towards skepticism about whether such approaches could be at all informative for discerning morphological homologies (Jenner 2006).
But this skepticism goes too far. As long as two or more approaches are targeted at the same overarching phenomenon (in this case evolutionary history of characters) it should at least be possible to take steps towards integrating their findings, ideally through methodological integration. In this particular context this means an understanding of morphological homology that takes into account evidence from various sources, including traditional morphological approaches as well as more recent molecular approaches: unity in the explanandum alongside diversity in the explanantia necessitates an integrative methodological framework. Here, the aim is to provide exactly that. I will bring into attention a recent model of homology: that of the Character Identity Mechanism or “ChIM” for short (DiFrisco et al. 2020). This lays the groundwork for such an integrative framework as is required for early metazoan research. I will then propose a three-level integrative approach based on this model that is centred on informing hypotheses of morphological homology by taking into account not only traditional morphological approaches but also developmental genetics, the degree of association between characters identified by each approach, and the interplay between statistical and causal explanations in a broader phylogenetic context. I will conclude with a brief discussion of prospects and challenges, as well as possible bearings on the philosophy of biology and philosophy of science more generally. But I will first go into a bit more depth on traditional and molecular approaches to homology and discuss their respective strengths and weaknesses in order to make a case for the importance of integrative approaches.
Homology, morphology, and phylogenetics: an incomplete picture
A brief history of homology
The term homology was coined by the 19th century naturalist Richard Owen in the following oft-quoted terms: “the same organ under every variety of form and function”. This “sameness” (Novick 2018; Ramsey and Siebels Peterson 2012) was deemed highly valuable as a concept that could enable the comparison of variants on the same fundamental “type” (organs or body parts) across different species, which in turn enabled robust taxonomic groupings such as vertebrates (vertebral column), tetrapods (four pentadactyl limbs), and so on (Hall 2013; Rieppel 1994). It also enabled comparisons between living and extinct organisms, and therefore the reconstruction of evolutionary history; though, in fact, while homology was initially conceived as an essentialist, ahistorical concept, the Darwinian view of evolution gave it a historical explanation on the grounds of common descent: under this view, homologues were traits that belonged to the “same” category because they had been inherited from a corresponding trait in the last common ancestor of the species exhibiting them (Hall 2012; Mayr 1982).
This evolutionary conception of homology was later solidified with the advent of cladistics and modern phylogenetics, where homologues formed the basis of phylogenetic classification as traits that were shared by two or more species; shared derived traits (synapomorphies) in particular, which pick out or define clades, were identified with homologies (Cracraft 2005; Patterson 1982, 1988). The value of this general approach became even more apparent with the rise of molecular genetics due to (1) the clear applicability of the homology concept to genes based on their patterns of inheritance and, as a consequence, their sequence similarities; and (2) the use of genetic homology to construct phylogenies based on genetic data that have, for the most part, corroborated phylogenies based on more traditional methods such as comparative morphology (Giribet and Edgecombe 2019; Nielsen 2012). Moreover, while homology was originally assessed on the basis of shape, transitional states, and topological correspondence of sub-parts (Remane 1956; Rieppel 1994), further criteria have since been added that are applicable to morphological as well as (arguably) genetic characters.
Nonetheless, phylogenetic/morphological approaches to homology struggle with addressing a number of conceptual issues. These include (1) character continuity, (2) serial homology, and (3) character individuation; and have recently been reviewed (DiFrisco, 2020). I will now briefly discuss these and the closely related issue of explaining homology.
Issues faced by the phylogenetic/morphological approach
One phylogenetic way to define homology is, as tacitly mentioned above, in terms of character continuity due to common descent (Hall 2012; Novick 2018; Wagner 2007, 2014). Under this view, the mouse Pax6 gene (a classic example) is homologous to the fly eyeless gene because they have been continuously present in both lineages since they split up sometime in the late Precambrian (Shubin et al. 2009). Likewise, the vertebrate lung and the teleost swim bladder are homologous due to the same reason. However, this appeal to continuity as something crucial to defining homology runs into two problems where morphological characters are concerned: the first is that unlike genes, morphological traits are in fact not normally continuously present since they need to develop anew in each generation; the second is that characters lost in evolution sometimes reappear (Collin and Miglietta 2008; Kohlsdorf and Wagner 2006; Kurtén 1963).
Another shortcoming of this general approach is that it fails to account for serial homologues. Broadly speaking, serial homologues consist of traits whose homologues are found within the same organism, rather than in other organisms in the same or different species (Hall 2012; Novick 2018). Examples include the serially repeated limbs of arthropods or their segments (repeated structures in general tend to be captured by this concept) at the morphological level, and paralogous rather than orthologous genes at the genetic level. The phylogenetic approach fails to account for serial homology because, in basic terms, the lineages along which serial homologues can be traced at best only partially overlap with species lineages—those with which phylogenetics is normally concerned (e.g. paralogous gene trees vs. species trees).
The third problem is the internal incompleteness of the phylogenetic approach to homology with respect to individuating traits: the phylogenetic approach on its own has no way of independently individuating—i.e. picking out—the characters whose homology it tests. These characters cannot be picked out at random: while some possible characters would be absurdly counterintuitive (e.g. eyebrows + appendix), even more obvious characters often need to be picked quite carefully in order to carry phylogenetic signal (e.g. whether the entire arm or the forearm should be taken as the individual character). Traditionally, phylogenetics has relied principally on comparative morphology (as well as embryology—see below) as the source of individuated characters, and this approach has been successful for the most part; and, in turn, better-resolved phylogenies resulting from better-individuated characters have informed character choice in comparative morphology (Hall 2012; Williams and Ebach 2020). However, there have been a number of notable cases where more recent molecular phylogenies (effectively independent evidence) have decisively invalidated previously widely-accepted character homologies by overturning long-held phylogenetic trees, leading to doubt about the sufficiency of morphological evidence alone in individuating characters. A notable example here is the breaking up of the former Articulata, which consisted of arthropods and annelids combined, based on the inference of homology between segmentation in arthropods and annelids among a majority of non-segmented protostome phyla (Garey 2001; Nielsen 2012).
The three issues briefly outlined here all point to a more general epistemic gap in the traditional approach to homology: they all result from an inherent shortcoming in providing causal explanations for character homology. What explains the reappearance of characters in ontogeny and occasionally phylogeny? What explains the intuitive sameness of serial homologues? Finally: what explains the individuation of characters? In other words: why do some characters exhibit ontogenetic and phylogenetic stability and individuality, whereas others do not?
Developmental genetics: conservation and dissociation
Developmental genetics
Morphological traits—at least stable, individuated ones—can be seen as consistent patterns that are caused by presumably consistent underlying processes. It is therefore plausible that in order to provide causal explanations for their ontogenetic and phylogenetic stability and individuality we should turn to investigating the stability and individuality of their underlying developmental processes. In this section, I will broadly outline one of the most prominent developmental approaches to homology—developmental genetics—and how it attempts to resolve the issues raised above. As we shall see, while this approach adds invaluable insight for the purposes of explaining homology, they have—at least so far—fallen short of achieving their purported aims for reasons I will discuss.
To start with, let us go back to the first problem: a purely phylogenetic account of homology has difficulty with the fact that morphological characters are generally not continuously present throughout ontogeny, and are in some cases not even continuously present throughout phylogeny (e.g. atavisms)Footnote 1.
One very abstract way to deal with this problem is to delegate homology to the level of the underlying information which is continuously present throughout ontogeny as well as phylogeny; in other words, to define homology in terms of “continuity of information” (Ramsey and Siebels Peterson 2012; Van Valen 1982). Somewhat more concretely, this can be referred to as the genetic level, since it is presumably the genes that are continuously present and contain the relevant information; in turn, this is because it is presumably the genes that control developmental processes. This forms the basis of the developmental genetic approach, whereby the homology of two or more characters is assessed (or even defined, depending on the account—see below) on the basis of the genes that are causally involved in its development. This is a highly functionalist and reductionist approach, in that it takes development to be a function of genes and how they interact with each other, the relevant information for all of which is taken to be contained within the genome of the organism; more specifically, it takes genes to fully explain traits—in other words, it reduces traits to genes (Griesemer 2001, 2006). This general approach also has the potential to resolve the other two problems raised in the previous section: serial homology and character individuation. By finding homology in the underlying genetic level, serial homologues can be recognised as such via their shared genetic underpinnings—which are of course expressed in a different morphogenetic context (e.g. forelimbs and hindlimbs). Similarly, the same key idea of genetic underpinnings of a trait could potentially capture traits that are (1) not arbitrarily defined because they are individuated on the basis of such genetic underpinnings, and which also are (2) phylogenetically and ontogenetically stable because their genetic underpinnings are phylogenetically and ontogenetically robust. Thus, pushing homology “down one level” seems, at first glance, to make it explainable in terms of the lower-level entities (genes) and their interactions.
Moreover, this approach has an obvious strength: genes are things that can be sequenced and experimentally manipulated, and the results of such experiments can be quantified and carefully examined, and it has thus been perceived as less “subjective” due to it being a task that is often not as straightforwardly carried out under the more traditional morphological methods. I will now turn to some rather unsurprising complications arising from this generally reductionist/functionalist approach.
Conservation and dissociation
Some of the most vital empirical findings supporting and fuelling a developmental genetic approach to homology have been the discoveries of highly conserved genes with highly conserved functional roles in the development of phylogenetically disparate animals (Carroll 1995; Davidson and Erwin 2006; Erwin and Davidson 2009; Wagner 2014); some of the most widely-known examples being the conserved role of Hox gene expression along the antero-posterior axis of bilaterians and the directive axis of anthozoans and its role in patterning the axes (Arendt 2018; Carroll 1995; Genikhovich and Technau 2017; He et al. 2018; Peterson et al. 2000), the conserved role of Pax6/eyeless in the formation of eyes in Drosophila and Mus (Halder et al. 1995), and the extensive role of conserved signalling pathways such as cWnt or cBMP in body plan and organ (especially limb) formation across bilaterians (DuBuc et al. 2018; Ferguson 1996; Genikhovich et al. 2015; Genikhovich and Technau 2017; Jager et al. 2013; Martindale 2005; Niehrs 2010; Nielsen et al. 2018; Panganiban et al. 1997; Shubin et al. 1997; Tarazona et al. 2019)—to name only a few. To be more explicit, the general argument from these cases is that the conservation of developmental genetic characters that underlie conserved morphological characters—or more succinctly the robustness of correlation/association between genetic and morphological traits—vindicates the developmental genetic approach as a potentially powerful explanatory tool for the stability and individuality of morphological traits and their homology.
Nevertheless, this issue was criticised quite early on by several authors on the basis that it does not always pick out the right homologues (Abouheif 1997; Bolker and Raff 1996; Dickinson 1995; Nielsen and Martinez 2003; Wray and Abouheif 1998). For example, morphological/phylogenetic approaches to homology decisively rule out the homology of vertebrate and insect eyes or that of bilaterian “limbs”; this has led to some authors questioning whether developmental genetics has anything at all to say about morphological homology (Jenner 2006). This leads us to a deeper question: where is the source of this tension between the two broad approaches? Why do developmental genetic characters tend often to be dissociated from/only partially associated with morphological characters, contrary to the assumptions of developmental genetics? Does this leave any space for developmental genetics being informative for inferring morphological homology?
First, let us examine the two broad causal processes that give rise to dissociation between morphological and developmental genetic characters over the course of phylogenesis. These are co-option and Developmental System Drift or DSD (Chipman 2010; Erwin, 2009; Haag 2014; Jaeger 2018; Lowe and Stolfi 2018; Nahmad et al. 2008; Sanetra et al. 2005; True and Haag 2001). Co-option is taken here to broadly refer to cases where the same (e.g. homologous) gene or set of genes has been recruited for a function other than the original function; a notable example is the co-option of the genetic system involved in limb formation in insects to produce butterfly “eyespots” (Brunetti et al. 2001). DSD broadly refers to (normally gradual) changes in the underlying genetics of a trait in spite of the stability of the trait itself—a well-documented evolutionary phenomenon most famously known from studies on the nematode vulva, where extensive DSD has occurred in relatively closely-related species (Haag 2014). In more abstract terms, of these two evolutionary mechanisms the robustness of association is broken—assuming the developmental genetic and morphological characters in question were associated to begin with (i.e. in the last common ancestor of the clade)—under one class of cases by the developmental genetic character remaining the same while the morphological character changes/is no longer the same (co-option) or vice versa (DSD).
Deep homology, developmental genetic definition of homology, and GRNs
There have been three general attempts at tackling the issue of dissociation without having to give up on the developmental genetic approach entirely. I will now briefly discuss these three attempts and argue that none of them are entirely satisfactory.
Deep homology
The concept of deep homology (Shubin et al. 1997, 2009) attempts to capture cases where two morphological traits that may or may not be homologous—e.g. vertebrate and arthropod eyes—have some shared underlying genetic basis such as the expression of Pax6/eyeless during their development. The claim that the two are deeply homologous is supposed to convey that both have likely evolved from a common eye progenitor, perhaps similar to the eyespots found in platyhelminths or acoels; and that co-option and/or DSD have resulted in the divergence of the morphological and the developmental genetic traits in question over the course of phylogenesis. By doing so, the concept has been taken to avert the conflict that results from dissociation between the two levels. However, this is a deeply unsatisfying approach: instead of explaining the individuation and stability of morphological traits such as eyes it explains them away by simply admitting that some traits have some features in common—e.g. some of the genes involved in their development—and thereby giving us no actual way of distinguishing between genuine morphological homology (e.g. eyes of insects and crustaceans) and convergence (e.g. eyes of vertebrates and insects). In other words, the deep homology of vertebrate and insect eyes has almost nothing to do with the homology of eyes present in adult organisms (i.e. whether the last common ancestor of vertebrates and insects had eyes comparable to the complex eyes present in the two groups), and almost everything to do with the presence of eye progenitors (i.e. simple “embryonic” eyes) in their last common ancestor.
Developmental genetic definition of homology
If one takes morphological homology as elusive or indeed “subjective”, leading to misguided phylogenetic trees and character reconstructions due to the inherent difficulties of discerning convergence from shared origins, then why not dispose of it altogether and come up with a new definition of homology based on the more quantifiable and “objective” genes? This is the approach that I have chosen to call the developmental genetic definition of homology. It consists of redefining homology as pertaining to genetic characters rather than morphological ones; in other words claiming that morphological homology is reducible to genetic homology. It ultimately relies on the fact that genes are highly amenable to homology claims due to their high-fidelity mode of inheritance but takes one big step further by claiming that genetic characters are, at least in practice, the only characters for which homology claims can reliably be made. This approach stems from studies on the evolution of cell types: though extensive gene expression data from various cell types—molecular fingerprints, in other words—were initially used as additional characters to assess cell type homology (Arendt 2008; Arendt et al. 2015, 2019), they were later taken to form a definition of cell type identity (and therefore homology) under the Core Regulatory Complex (CoRC) model (Arendt et al. 2016). This is unsurprising because in terms of levels of organisation cells are not far above genes, and it is therefore plausible that their properties could be to some extent reducible to those of their genes. Nonetheless, by shifting the focus from morphological to genetic characters this approach misses the explanandum—which is, again, the individuation and stability of morphological traits—in a manner similar to but stronger than deep homology: in other words, its explanations cease to be about what was originally meant to be explained. Thus, while it might be a useful methodological commitment to deal with questions about cell type homology, it is less likely to be useful for approaching the homology of higher-level traits such as organs and organ systems.
Gene regulatory networks
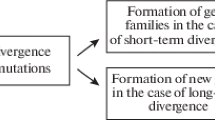
A more overarching approach—indeed virtually present in most rigorous developmental genetic studies (Wagner 2014) (including those briefly discussed in the preceding paragraphs)—consists of not focusing simply on one or a few genes but sets of genes and how they functionally interact with each other in a Gene Regulatory Network or GRN (Davidson and Erwin 2006). GRNs are potentially far more explanatory because virtually no morphological trait is underpinned by the expression of one or a few genesFootnote 2; this is especially true of complex traits such as organs which are of most interest to comparative research. Crucially, the relative complexity of GRNs makes them potentially far more useful in tracing homology and discerning it from convergence. Thus, it is far more plausible that if morphological homology is to be traced by proxy of some underlying genetic factors, these will come in the form of a GRN. Indeed, one of the most sophisticated and demonstrably explanatory developmental genetic models of homology is that of Character Identity Networks (ChINs) (Wagner 2007, 2014), where a ChIN is a conserved GRN that underlies the identity and individuality of stable morphological traits such as insect forewings, or metazoan cell typesFootnote 3 (Fig. 1). The ChIN model attempts to resolve the issue of conservation and dissociation via a three-stage model of the genetics of character development. In the first stage, positional information is encoded by signalling gradients resulting in the determination of character position (e.g. position of the forewing in the body plan); in the second stage, the ChIN is expressed and determines the identity of the developing morphological trait (e.g. forewing rather than hindwing or antenna); and in the last stage “realiser/effector” genes determine the state of the morphological character (e.g. elytron rather than flight wing). Under this model, while all three stages are causally involved in the generation of the morphological character, only the second stage needs to be conserved as it is the stage that determines the identity of the character (two characters with the same identity would be considered homologous). In fact, this model fits nicely with an abundance of evidence (Wagner 2014). Evolutionary changes in the “upstream” positional information genes are irrelevant to homology as long as the same character develops (this also neatly explains serial homology), and changes in the “downstream” realiser only result in changes in the character state (elytra vs flight wing) and not changes in character identity (forewing rather than hindwing).
Character Identity Mechanisms are evolutionarily stable relative to upstream signals (positional information) and downstream effectors (realiser genes), as represented by the hourglass figure; modified from (DiFrisco et al. 2020)
Nevertheless, the ChIN model has some serious limitations: while ChINs are envisaged as networks of transcription factors, some key metazoan characters are determined by signalling pathways—most notably the basic eumetazoan body plan (see below). Furthermore, it does not address the role of other regulatory factors such as non-coding RNAs in character identity. It therefore seems that the ChIN model is only explanatory in a limited number of cases, and a more general model of character identity is needed in order to expand its explanatory scope. This is the topic of the next section.
Character identity mechanisms: a new model of homology
The ChIN model has been modified in a recent paper (DiFrisco et al. 2020) primarily in order to overcome the problem of its limited explanatory scope. The new model is termed Character Identity Mechanisms (ChIMs). As the name suggests, the use of the word mechanism is intended to capture a broad class of phenomena that includes ChINs, but extends much further by not being restricted to GRNs as the sort of things that could in principle explain the individuation and stability of morphological traits; rather, the mechanisms now comprise processes that underlie phenotypic traits at different levels of organisation including cell types, simple and complex tissues, and organs—each of these requiring their own class of developmental processes. A distinguishing feature of the ChIM model is that it defines ChIMs, roughly speaking, as processes that possess a number of core properties, though none of these properties are neither necessary nor sufficient. Rather, they form a property cluster which means they tend to be causally interconnected and therefore frequently co-occur. These include modularity, complex organisation, necessity for particular functional outcomes, causal non-redundancy, less replaceability compared to upstream signals and downstream effectors, a high degree of burden, and high evolutionary conservation, which together should give rise to phylogenetic traceability. Collectively, what these properties achieve is precisely what is needed for an explanatory model of homology: they enable ChIMs to explain the stability and individuation of morphological characters by virtue of their own stability and individuation, both ontogenetically and phylogenetically, without attempting to reduce morphological traits to underlying mechanisms. Furthermore, having a cluster of properties allows the incorporation of a broad range of causally distinct processes underlying distinct morphological traits (e.g. cell types and organs).
Of special importance in the context of inferring homology is traceability. One of the key criteria for inferring homology is that the trait under study should be as traceable in phylogeny as possible (though this is often hard to establish in practice), since traceability enables the researcher to discern cases of homology from cases of homoplasy where traits appear independently in phylogeny and are therefore not continuously present down to the last common ancestor of the species, and are thus ontologically untraceableFootnote 4. Traceability at the level of underlying mechanisms can further corroborate hypotheses of complexity where morphological evidence alone is insufficient to infer homology or non-homology.
Developmental mechanisms and morphological homology: an integrative approach
Perhaps the most attractive feature of the ChIM model for early metazoan research is that it lays the groundwork for a truly integrative approach to inferring morphological homology, something that is painfully absent from the existing literature. The aim of this section is to introduce exactly such an approach. As we will see, this approach can account for evidence from comparative morphology as well as developmental biology (including comparative embryology and developmental genetics) and integrate them using a combination of statistical/correlational and mechanistic/causal methods, all within an overarching phylogenetic background.
Inferring trees and inferring homologies
As a prelude, it should be stressed that there are two broad classes of phylogenetic inferences both interwoven with homology inferences: (1) inferring the topology of phylogenetic trees(i.e. the way lineages are related to each other), and (2) inferring the characters and character states of ancestral species. In the former class character homologies are held fixed by being inferred independently, for example through classical methods such as anatomical correspondence or comparison with fossils in the case of morphological traits, sequence similarity in the case of genes, and the four “handles for homology” in the case of ChIMs. The distribution of homologous characters is then used to infer the most plausible explanation in terms of tree topologyFootnote 5, normally via Bayesian, maximum likelihood, or parsimony-based methods. Nonetheless, tree topology can be informative for inferring character homologies, especially for characters of uncertain homology status. Consider the revision of homology between arthropod and annelid segmentation (Garey 2001) or the shared lack of a coelom in platyhelminths and acoels (Hejnol and Pang 2016) in light of more recent molecular phylogenies. This brings us to the latter class of inferences, whereby character homologies—and thus the characters and character states of ancestral species—are inferred against the ideally fixed background of tree topologies.
Crucially, this shows us that in order to infer character homologies it is never sufficient to rely exclusively on phylogenies (whether using congruence-based methods of the very same characters or independent evidence) or evidence pertaining to the characters themselves (e.g. anatomical correspondence). The best possible course of action is to find the most plausible evolutionary scenarios using all available evidence, though this often means breaking down our scientific questions into classes such as the two distinguished here, which would require idealisations such as assuming a fixed tree topology. Indeed, the integrative approach to be discussed now makes this exact idealisation in order to retain explanatory power; though, as we shall see near the end of this section, it can be further integrated into a more overarching framework incorporating methods aimed at inferring tree topologies.
Correlational and mechanistic approaches
So far, we have seen how developmental approaches to homology rest on the assumption that if the mechanisms underlying morphological/phenotypic characters (be they genes, morphogenetic processes, etc.) are understood then one can make more precise inferences about the homology of these characters; and that this tends to run into problems because such underlying mechanisms are often dissociated to lesser or greater degrees from those characters across phylogeny. Often the problem has been that what was initially identified as the mechanism later turned out to be merely a non-essential part of the mechanism, such as specific genes involved in the development of vertebrate and insect eyes (Shubin et al. 2009) or nematode vulvas (Haag 2014). This problem can be construed in two ways: firstly, as an error in identifying the right mechanism; and, secondly, as a mismeasurement of the degree of association between the morphological and developmental characters. The first construal is relevant to causal/mechanistic methods: it opens the way for experimental science to attempt to identify the right mechanism, and the ChIM model is the prime conceptual candidate to guide this type of research. The second construal is relevant to correlational/statistical methods: it opens the way for the measurement of degrees of association between characters of the two kinds, which in turn enables the integration of developmental evidence into a phylogenetic approach aimed at inferring homologies. Furthermore, such a method would need to rest on a conceptual basis that provides guidelines on how to make inferences based on the evidence.
A key conceptual requirement for evidence to be useful for making homology inferences consists in satisfying the following three criteria: effectiveness, admissibility, and informativity. I will now explain these in turn using examples from the literature, which will simultaneously elucidate how developmental evidence can be integrated into a phylogenetic approach. Next, I will turn to the more overarching integrative approach to inferring homology.
Effectiveness
Effectiveness of developmental evidence in this context refers to the degree to which it can potentially change prior confidence in homology or non-homology of a morphological trait. Consider the classic example of the non-homology of vertebrate and insect eyes. In this and other similar cases, morphological analysis strongly disconfirms the hypothesis of homology between these two organs: both have highly complex structures with barely any anatomical correspondence of parts between them and other living bilaterian lineages and the fossil record shows clearly that the organs cannot be traced back to their last common ancestor (Nielsen 2012). Therefore, even relatively strong developmental genetic evidence—e.g. similarities between GRN structure rather than mere shared use of specific genes—should point to co-option rather than homology: the developmental evidence here is therefore dubbed as ineffective. The same could be said about cases where morphological studies are conclusive with respect to character homology, such as the aforementioned nematode vulva: differences in underlying genetics are best interpreted as being a result of DSD rather than morphological convergence (Haag 2014). It is only in cases where morphological evidence alone is inconclusive that developmental evidence (as well as other sources of evidence more generally) can potentially tilt the inference in favour of either homology or non-homology—thereby being effective.
Admissibility
In order for developmental evidence to be admissible, the developmental characters themselves need to be homologous across the clade under study. Genuine cases of inadmissibility are difficult to point out because virtually any study seeking to use developmental evidence starts with the discovery of conserved underlying genes or GRNs. Nonetheless, admissibility is indeed a requirement for the validity of integrative homology inferences: suppose two genes deemed previously to be orthologous underlying the same morphological trait turned out to be in fact paralogous. Since paralogous genes are frequently co-opted to perform similar functions, the evidence would be less admissible than previously assumed. The importance of admissibility should become clearer when it is put together with effectiveness to feed into the next criterion: informativity.
Informativity
Homologous genes, GRNs, or morphogenetic processes—i.e. developmental traits—combined with uncertainty from morphology alone has nothing to say about morphological homology. As discussed above, the final ingredient is the degree of association between characters at the two levels. Consider a relatively recent study (Martín-Durán et al. 2018) where the authors argue for the convergent evolution of bilaterian nerve cords. In their analysis, they first show that the common expression pattern of a number of homeobox genes involved in the spatial specification of nerve cords in a few classic model organisms is in fact not shared by representatives from taxa that had not been previously studied, which nonetheless possess nerve cords. They then cite this as a reason against the informativity of the developmental genetic evidence in this case, followed by an inference of non-homology of the nerve cords across Bilateria based on morphological evidence alone. In other words, the authors show that there is no significant degree of association between characters at the two levels, and the evidence is therefore uninformative.
Granted that the developmental evidence provided is both effective and admissible, a strongly positive and a strongly negative association are both informative: the former suggests homology, and the latter non-homology. While I am unaware of any examples of the latter (possibly due to selection bias), the next part of this paper (Reconstructing the ancestral eumetazoan body plan) will provide a strong example of the former.
Three levels of integration
Before turning to the case of the eumetazoan body plan, it should be emphasised that the overarching framework being proposed here consists of three levels of evidential integration. These are (1) integrating developmental and morphological evidence in a correlational approach, (2) integrating the correlational approach with a causal/mechanistic approach to infer homology more precisely, and (3) integrating homology inferences with phylogenetic tree inferences to come to evolutionary scenarios that account for as many sources of evidence as fully as possible. I will now discuss the first and second levels in the following paragraphs but forego discussion of the third in this paper for want of space.
As we saw in the preceding subsection, the first level of integration basically consists in finding correlations between morphological and developmental characters and using such correlations and, if the relevant criteria are met, to update previously uncertain homology inferences (Fig. 2). The main reason why it is merely correlations between characters that are being utilised here is that degrees of correlation can feature in the essentially statistical science of cladistic phylogenetics. Put another way, the purpose of this correlational approach is to estimate the probability of the presence/absence of a character in the last common ancestor of a clade—to estimate the probability of character homologyFootnote 6. The core features of this approach can be conceived in Bayesian terms where the prior is the probability of character homology given morphological evidence alone, and the posterior is the probability of character homology given morphological and developmental evidence combined. The more strongly the criteria described above are met (especially informativity), the more the posterior is affected by the developmental evidence.
Effectiveness, admissibility, and informativity of developmental evidence when integrated with morphological evidence. The x axis represents confidence in developmental genetic homology, the y axis represents confidence in morphological homology based on morphological evidence alone, and the z axis represents degree of association (= correlation) between the two traits across a clade. All three parameters range (-1, 1)
Nevertheless, correlations alone can only provide partial explanations of complex causal phenomena: “correlation does not imply causation”. In this particular context, it is crucial to recognise that correlations between developmental and morphological characters, no matter how strong they might be, cannot be taken as conclusive evidence for a causal relationship between the characters because they could simply be spurious. Another shortcoming of this approach that also comes down to its inherent non-causality is its weakness in identifying candidate developmental characters, be they genes, GRNs, or signalling pathways. It is for these reasons that this approach must always be complemented with a causal/mechanistic approach. As argued earlier, the best candidate approach of this kind is the ChIM model discussed above: it allows more precise identification of candidate developmental mechanisms underlying morphological characters being studied; and it simultaneously provides causal explanations of the relations between the characters, thereby minimising the risk of making inferences based on spurious correlations. Thus, the integration of the correlational approach with a causal/mechanistic approach is necessary for making robust homology inferences (Fig. 3), which in turn both inform and are informed by tree inferences to arrive at integrative evolutionary scenarios of biological evolution (Nejad Kourki 2021).
An algorithmic model of integrating developmental and morphological evidence, corresponding to the first, correlational level of integration and the application of the three criteria. The last step in the process forms the basis of the second, causal/mechanistic level of integration: as homology is suggested by the correlational analysis, it can be further confirmed or refuted by experimental evidence, which can then feed into further correlational analysis
Reconstructing the ancestral eumetazoan body plan
In this part of the paper, I will use the example of reconstructing the ancestral eumetazoan body plan as a case where the integrative approach outlined above is highly applicable. In doing so, I will both utilise the ChIM approach as well as arguing that the eumetazoan body plan ought to be added to the set of phenomena whose homology this approach purports to explain (i.e. cell types, simple and complex tissues, and organs).
Old debate, limited evidence
The possibility of homology between eumetazoan body plans was recognised as early as the early 1800s (Hall 2012), even before Darwin or the precisification of biological sameness into the concept of homology. The most notable example of an early evolutionary scenario linking together the basic body plans of the various metazoan taxa, however, is to be found in the works of Haeckel who formulated the Blastaea/Gastraea hypothesis (Nielsen 2012; Nielsen et al. 2018). According to this hypothesis—which has survived into modern times in one form or another—the earliest animals (the Blastaea stage) had the general body plan of a blastula, and later (the Gastraea stage) that of a gastrula. What needs to be emphasised here is that inferring the Blastaea and Gastraea stages in phylogeny—i.e. reconstructing ancestral metazoans as such—relies virtually exclusively on observations of ontogeny—the developmental stages of living animals (the blastula and the gastrula stages). This is the paradigmatic case of applying Haeckel’s dictum: “ontogeny recapitulates phylogeny”.
Nevertheless, evidence accrued since Haeckel’s days does not support universal application of his dictumFootnote 7. Furthermore, the historical overreliance on comparative embryology to elucidate early metazoan evolution stems in large part from the historical absence of virtually any other source of evidence to fuel research in the area. This has changed in recent times by discoveries of fossils from the early Cambrian as well as the late-Precambrian (Budd 2015) and advances in analysis and interpretations of these fossils; as well as the use of developmental genetic methods to discover homology relations between characters found in animals belonging to disparate taxa. As the impact of modern palaeontology on this debate is beyond the scope of this paper, I will only discuss the impact of developmental genetics, especially in relation to the ChIM model and the integrative approach.
The mother of all organs: the ancestral eumetazoan body
The ancestral eumetazoan body plan
The clade Eumetazoa is comprised by at least the two major clades Bilateria and Cnidaria and, somewhat more contentiously, the Ctenophora (Dunn et al. 2008; Laumer et al. 2019; Pisani et al. 2015; Zhao et al. 2019); the only living non-eumetazoan animals are the sponges (Porifera) and Placozoa. Given the uncertain phylogenetic placement of ctenophores and the uncertainty about the homology of their body plans to those of bilaterians and cnidarians, I will only focus on the latter two here. Bilateria includes the majority of animal phyla, such as the well-known chordates, arthropods, molluscs, annelids, and flatworms; as well as the somewhat obscure kinorhynchs, hemichordates, loriciferans, and priapulids—just to name a few (Nielsen 2019). Cnidarians are subdivided into two major clades: Anthozoa and Medusozoa. Sea anemones and numerous species of corals belong to Anthozoa, while jellyfish, other corals, the classic model organism Hydra, and the notorious Portuguese man-of-war belong to Medusozoa.
Even though there is a vast diversity of derived body plans to be found across the bilaterian and cnidarian phyla, virtually all of them share a conserved stage in their early development: the gastrula. Furthermore, both anthozoans and bilaterians exhibit bilateral symmetry, and recent evidence suggests that bilateral symmetry is in fact ancestral to the last common ancestor of Cnidaria and Bilateria (DuBuc et al. 2018; Genikhovich et al. 2015; Genikhovich and Technau 2017; Nielsen et al. 2018); this implies secondary radialisation of medusozoans—supported by their highly derived genomes (Gold et al. 2019). Crucially, developmental genetics reveals the conserved utilisation of specific signalling pathways—namely the canonical Wnt and canonical BMP pathways (Genikhovich et al. 2015; Genikhovich and Technau 2017; Niehrs 2010) in the spatial specification of the gastrula stage. The interactions between the two signalling centres and their associated signal/ligand gradients gives rise to a three-dimensional coordinate system which in turn underlies the formation of a bilateral body plan through the movement of cell layers and differentiation of tissues in the embryo (DuBuc et al. 2018; Eivers et al. 2009; Niehrs 2010; Wijesena et al. 2017), aided by the conserved spatiotemporal expression of transcription factors, especially homeobox genes, which function in patterning along the resulting body axes.
The origin of organ ChIMs
The utilisation of conserved signalling pathways linked with conserved spatiotemporal expression of transcription factors features in the ChIM model as constituting the kind of ChIM that underlies organs (DiFrisco et al. 2020). A paradigmatic example is the development of the vertebrate limb, which utilises the BMP and Wnt signalling pathways as well as additional ones such as FGF signalling and the expression of homeobox genes to achieve a three-dimensional organisation strongly reminiscent of the eumetazoan body plan itself (Fig. 4). The similarities between organs and the body plan do not end here. While both are conserved both at the morphological level and at the ChIM level, thereby constituting a character with an individuated identity, they also both vary with respect to their character states: in the case of vertebrate limbs (and similarly the highly similar but most likely convergent arthropod limbs) these include fins, arms, legs, wings, etc.; in the case of the body plan, these include the diverse phylotypic stages of various bilaterians as well as the somewhat divergent anthozoan body plan (discussed below). In further accordance with the ChIM model, the developmental processes upstream of the gastrula stage is highly variable: the exact process of gastrulation is highly clade-dependent and ranges from the relatively straightforward invagination in numerous taxa to the highly derived and complex ingression.
Interaction between Wnt and BMP signalling gradients in the early embryo of a cnidarian, giving rise to a bilateral body plan in later stages, representing (as a proxy) the ancestral eumetazoan; modified from (Genikhovich and Technau 2017)
The significance of this deep similarity between the eumetazoan body plan and a paradigmatic bilaterian organ (the limb) is that it strongly suggests the co-option of the gastrula ChIM in the emergence of bilaterian organs—at least the vertebrate and arthropod limbs but possibly other organs which utilise similar signalling pathways in a conserved spatiotemporal manner. This in turns allows the explanation of the emergence of eumetazoan (especially bilaterian) complexity in terms of the emergence and repeated co-option and subsequent divergence of conserved mechanisms present in the last eumetazoan common ancestor, as well as providing an example of the explanatory value of the ChIM model in the case of early animal evolution, expanding the original set of explananda (cell types, tissues, and organs). By doing so, it also paves the way for testing more specific hypotheses surrounding the evolution of the various eumetazoan body plans. I will now describe one such hypothesisFootnote 8.
Which end is which: the integrative approach to axis homology
The eumetazoan body plan is a character identity that comes in variable states. Notably, the homology relations between the bilaterian and anthozoan body axes are still unresolved, leaving room for effective developmental evidence: according to one hypothesis, the oral-aboral (OA) axis of Anthozoa (as well as Medusozoa and Ctenophora) is homologous to the bilaterian antero-posterior (AP) axis, and the anthozoan directive axis is homologous to the bilaterian dorso-ventral (DV) axis. This hypothesis rests primarily on the position of the blastopore and the extension of the gastric cavity/canal along the OA axis in Cnidaria and the AP axis in Bilateria. It is furthermore supported at the developmental genetic level by the shared utilisation of the cWnt pathway in specifying the OA and AP axes, and the cBMP pathway in specifying the directive and DV axes (Genikhovich and Technau 2017; Niehrs 2010)—both amounting to homology of developmental characters and therefore admissibility of the developmental evidence.
The alternative hypothesis is that the cnidarian OA axis is in fact homologous to the bilaterian DV axis, and the bilaterian AP axis is homologous to the anthozoan directive axis (Genikhovich and Technau 2017; Nielsen et al. 2018) (recall that medusozoans lack—and have most likely secondarily lost—the directive axis and are therefore radially symmetric). Based on morphological evidence alone, this hypothesis is harder to defend because it implies the detachment of the site of gastrulation/position of the blastopore/direction of gastric cavity from the DV axis in bilaterians. Regardless, this is precisely what developmental genetic evidence suggests: the cWnt signalling maximum is strongly associated with the site of gastrulation, which is in fact quite variable across Bilateria—sometimes posterior, sometimes anterior, and sometimes ventral—and is therefore not directly informative for inferring the homology of body axes. The cBMP signalling gradient invariably lies perpendicular to the cWnt signalling gradient across bilaterians and anthozoans studied so far probably due to their joint role in specifying a three-dimensional bilaterian body plan and is therefore only as informative as the cWnt signalling. Nevertheless, the linear expression of homeobox genes is conserved across both clades (and therefore admissible) and is strongly associated with the bilaterian AP axis as well as the anthozoan directive axis, making it the only informative developmental genetic character for inferring the homology of body axes between the two clades.
Thus, while clear empirical evidence is needed for the substantiation of this hypothesis, viewing it in light of the integrative framework proposed in this paper provides a clear way in which such evidence can be utilised for inferring the homology of body axes between Bilateria and Anthozoa. With respect to the three levels of integration, the example used here corresponds mainly to the first, correlational level: evidence is first shown to be potentially effective, then admissible, and then informative. Though the sketch provided here can certainly be enhanced by empirical data and statistical analyses, it does point towards and pave the way for the identification of developmental characters underlying the eumetazoan body plan corresponding to the second, causal/mechanistic level of integration.
Conclusions
To summarise, we have seen how over-emphasising the importance of approaches leaning too strongly towards either pure comparative morphology or developmental genetics is counter-productive to inferring homology relations and by consequence constructing evolutionary scenarios that most accurately reflect the actual history of life on Earth; and, using the example of the eumetazoan body plan, how an encompassing approach that integrates evidence and methodology at the three levels of analysis discussed above can resolve this issue, in large part by relying on the best mechanistic approach to homology proposed so far—namely the ChIM model. It would be interesting to see if a more generalised integrative approach of this kind—i.e. one that algorithmically progresses from less to more inclusive levels of analysis—could be potentially useful in answering questions such as how to integrate molecular and morphological phylogenies and even linguistic and biological phylogenies in the case of recent human evolution. Such an approach could start with assessing one kind of evidence independently (e.g. morphological), and then attempt integration of the kind suggested here with another relevant kind of evidence (e.g. developmental genetic) and move on to integrate the result (e.g. character homology) with another kind of evidence (e.g. molecular phylogenetics) to arrive at a more inclusive scenario (e.g. pattern of evolution of character across clades). It would also be interesting to see whether it could be applicable even outside of evolutionary science and whether it might have any bearing on the broader philosophy of science.
This is not to say that this approach and the ChIM model on which it relies are free from limitations. Most notably, in their current form they do not make any strong attempt to incorporate relevant extrinsic factors such as ecological pressures and certain intrinsic factors such as physico-chemical constraints on characters in the study of homology, and the ChIM model has difficulty capturing morphological traits that do not obviously have a ChIM and are rather “by-products” of other ChIMs, such as the human chin (DiFrisco, 2020; DiFrisco et al. 2020)—though this criticism could be avoided by claiming that these are not proper characters, as per Lewontin and Gould’s original argument (Gould and Lewontin 1979). Nonetheless, the ChIM model has notable merits besides those discussed above, including retaining the explanatory strengths of the ChIN model and extending its breadth of scope, as well as providing more than one “handle for homology”—which means their homology (not homology of the characters they underlie) can be assessed by several means, which allows for utilising different such means in different situations. Perhaps most importantly to the philosophy of biology and more generally philosophy of science, the ChIM approach is strictly a model rather than an account of homology, thereby effectively avoiding the decades-old quarrel over what homology is and instead focusing on how it could be explained under various circumstances. My hope is that this powerful approach can be efficiently integrated with phylogenetic practice under the model proposed in this paper.
Finally, it is worth noting that much of the confusion surrounding the relation between morphological traits and developmental genetic traits, often reflected in the idea of a “genotype-phenotype map” (Stadler and Stadler 2006), likely stems from a philosophical outlook that I call “reductionist functionalism”, inspired by Griesemer (Griesemer 2001, 2006). Functionalism in this sense roughly refers to an emphasis on the function of genes in organismal development, which often leads to a kind of reductionism whereby genes are taken to be the principal—if not the sole—bearers of biological explanation, as can be found in the works of Dawkins (Dawkins 1982, 2016), among others. The antidote to this general outlook might well lie in the recognition of the autonomy of morphological characters—i.e. recognising their quasi-independent identities and evolutionary trajectories that enable them to be definable without necessary recourse to underlying levels—and the often highly complex developmental processes underlying them, as well as other relevant factors such as those mentioned in the previous paragraph. The integrative approach outlined in this paper seeks to achieve precisely such a recognition in the hopes of a more holistic, perspectivalist, and overall organismic approach to the study of the history of life on Earth.
Availability of data and material
N/A.
Code Availability
N/A.
Notes
It should be noted that while the occasional lack of phylogenetic continuity seems genuinely problematic for an account of homology that relies on sameness due to phylogenetic continuity, the general lack of ontogenetic continuity, even though sometimes raised a serious issue (Novick 2018), in fact is not so. This is because a phylogenetic account ought to be insensitive to continuity (or lack thereof) on ontogenetic timescales. It is irrelevant to know how characters appear in each generation of each species, as long as they are in fact normally present within given species at the relevant point in the life cycle (Wagner 2014).
Notwithstanding traits with simple inheritance—the “Mendelian” traits”. In these cases as well as more generally, single genes never really “underlie” a trait in a strong sense; rather, they act as “difference makers” (Schäfer 2003).
This is in reference to the CoRC (Core Regulatory Complex) model of cell type specification. While this model “comes with” a developmental genetic definition of homology its explanatory power is not diminished by the definition as they are in fact logically independent of each other. For more details see (DiFrisco and Jaeger 2020).
Epistemological untraceability would simply result from lack of evidence and is not the issue at stake here.
Strictly speaking, tree topology is merely a formal representation of the historical branching of lineages within a cladistic model.
Probability of character homology and probability of the presence/absence of a character in the last common ancestor of a clade are only identical under phylogenetic concepts of homology (Cracraft 2005; Patterson 1988). Since they are presented here in a phylogenetic context, it is perfectly sensible to pragmatically identify one with the other.
Though under certain conditions the dictum does hold: specifically, where the developmental stages are serially causally dependent on one another. A notable example where this holds is Richard Prum’s model of feather identity and origin (Prum and Brush 2002).
It is worth noting that the homology of the gastrula stage does not imply the existence of a “Gastraea” stage in evolution—only that the last common eumetazoan ancestor also had a gastrula stage in its ontogeny, which is distinct from it corresponding to the adult form of this organism: Haeckel’s dictum is not vindicated.
References
Abouheif E (1997) Developmental genetics and homology: a hierarchical approach. Trends Ecol Evol 12(10):405–408
Arendt D (2008) The evolution of cell types in animals: emerging principles from molecular studies. Nat Rev Genet 9(11):868–882
Arendt D (2018) Hox genes and body segmentation. Science 361(6409):1310–1311
Arendt D, Benito-Gutierrez E, Brunet T, Marlow H (2015) Gastric pouches and the mucociliary sole: setting the stage for nervous system evolution. Philosophical Trans Royal Soc B: Biol Sci 370(1684):20150286
Arendt D, Bertucci PY, Achim K, Musser JM (2019) Evolution of neuronal types and families. Curr Opin Neurobiol 56:144–152
Arendt D, Musser JM, Baker CV, Bergman A, Cepko C, Erwin DH, Laubichler MD (2016) The origin and evolution of cell types. Nat Rev Genet 17(12):744–757
Bolker JA, Raff RA (1996) Developmental genetics and traditional homology. BioEssays 18(6):489–494
Brunetti CR, Selegue JE, Monteiro A, French V, Brakefield PM, Carroll SB (2001) The generation and diversification of butterfly eyespot color patterns. Curr Biol 11(20):1578–1585
Budd GE (2015) Early animal evolution and the origins of nervous systems. Philosophical Trans Royal Soc B: Biol Sci 370(1684):20150037
Carroll SB (1995) Homeotic genes and the evolution of arthropods and chordates. Nature 376(6540):479–485
Chipman AD (2010) Parallel evolution of segmentation by co-option of ancestral gene regulatory networks. BioEssays 32(1):60–70
Collin R, Miglietta MP (2008) Reversing opinions on Dollo’s Law. Trends Ecol Evol 23(11):602–609
Cracraft J (2005) Phylogeny and evo-devo: characters, homology, and the historical analysis of the evolution of development. Zoology 108(4):345–356
Davidson EH, Erwin DH (2006) Gene regulatory networks and the evolution of animal body plans. Science 311(5762):796–800
Dawkins R (1982) The extended phenotype, vol 8. Oxford University Press Oxford
Dawkins R (2016) The selfish gene. Oxford university press
Dickinson W (1995) Molecules and morphology: where’s the homology? Trends Genet 11(4):119–121
DiFrisco J (2020) Toward a Theory of Homology: Development and the De-Coupling of Morphological and Molecular Evolution. British Journal for the Philosophy of Science
DiFrisco J, Jaeger J (2020) Genetic causation in complex regulatory systems: an integrative dynamic perspective. BioEssays 42(6):1900226
DiFrisco J, Love AC, Wagner GP (2020) Character identity mechanisms: a conceptual model for comparative-mechanistic biology. Biology & Philosophy 35(4):1–32
DuBuc TQ, Stephenson TB, Rock AQ, Martindale MQ (2018) Hox and Wnt pattern the primary body axis of an anthozoan cnidarian before gastrulation. Nat Commun 9(1):1–12
Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Edgecombe GD (2008) Broad phylogenomic sampling improves resolution of the animal tree of life. Nature 452(7188):745–749
Eivers E, Demagny H, De Robertis E (2009) Integration of BMP and Wnt signaling via vertebrate Smad1/5/8 and Drosophila Mad. Cytokine Growth Factor Rev 20(5–6):357–365
Erwin DH (2009) Early origin of the bilaterian developmental toolkit. Philosophical Trans Royal Soc B: Biol Sci 364(1527):2253–2261
Erwin DH, Davidson EH (2009) The evolution of hierarchical gene regulatory networks. Nat Rev Genet 10(2):141–148
Ferguson EL (1996) Conservation of dorsal-ventral patterning in arthropods and chordates. Curr Opin Genet Dev 6(4):424–431
Garey JR (2001) Ecdysozoa: the relationship between Cycloneuralia and Panarthropoda. Zoologischer Anzeiger-A Journal of Comparative Zoology 240(3–4):321–330
Genikhovich G, Fried P, Prünster MM, Schinko JB, Gilles AF, Fredman D, Technau U (2015) Axis patterning by BMPs: cnidarian network reveals evolutionary constraints. Cell Rep 10(10):1646–1654
Genikhovich G, Technau U (2017) On the evolution of bilaterality. Development 144(19):3392–3404
Giribet G, Edgecombe GD (2019) “Perspectives in Animal Phylogeny and Evolution”: A decade later. In: University of Padova Press
Gold DA, Katsuki T, Li Y, Yan X, Regulski M, Ibberson D, Greenspan RJ (2019) The genome of the jellyfish Aurelia and the evolution of animal complexity. Nat Ecol Evol 3(1):96–104
Gould SJ, Lewontin RC (1979) The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proceedings of the royal society of London. Series B. Biological Sciences, 205(1161), 581–598
Griesemer J (2001) The units of evolutionary transition. Selection 1(1–3):67–80
Griesemer J (2006) 8 Genetics from an Evolutionary Process Perspective. Genes in development. Duke University Press, pp 199–237
Haag ES (2014) The same but different: worms reveal the pervasiveness of developmental system drift. PLoS Genet 10(2):e1004150
Halder G, Callaerts P, Gehring WJ (1995) New perspectives on eye evolution. Curr Opin Genet Dev 5(5):602–609
Hall BK (2012) Homology: The hierarchial basis of comparative biology. Academic Press
Hall BK (2013) Homology, homoplasy, novelty, and behavior. Dev Psychobiol 55(1):4–12
He S, Del Viso F, Chen C-Y, Ikmi A, Kroesen AE, Gibson MC (2018) An axial Hox code controls tissue segmentation and body patterning in Nematostella vectensis. Science 361(6409):1377–1380
Hejnol A, Pang K (2016) Xenacoelomorpha’s significance for understanding bilaterian evolution. Curr Opin Genet Dev 39:48–54
Jaeger J (2018) Shift happens: the developmental and evolutionary dynamics of the gap gene system. Curr Opin Syst Biology 11:65–73
Jager M, Dayraud C, Mialot A, Queinnec E, Le Guyader H, Manuel M (2013) Evidence for involvement of Wnt signalling in body polarities, cell proliferation, and the neuro-sensory system in an adult ctenophore. PloS one, 8(12)
Jenner RA (2006) Unburdening evo-devo: ancestral attractions, model organisms, and basal baloney. Dev Genes Evol 216(7–8):385–394
Kohlsdorf T, Wagner GP (2006) Evidence for the reversibility of digit loss: a phylogenetic study of limb evolution in Bachia (Gymnophthalmidae: Squamata). Evolution 60(9):1896–1912
Kurtén B (1963) Return of a lost structure in the evolution of the felid dentition. Societas Scientiarum Fennica
Laumer CE, Fernández R, Lemer S, Combosch D, Kocot KM, Riesgo A, Giribet G (2019) Revisiting metazoan phylogeny with genomic sampling of all phyla. Proceedings of the Royal Society B, 286(1906), 20190831
Lowe EK, Stolfi A (2018) Developmental system drift in motor ganglion patterning between distantly related tunicates. EvoDevo 9(1):1–15
Martín-Durán JM, Pang K, Børve A, Lê HS, Furu A, Cannon JT, Hejnol A (2018) Convergent evolution of bilaterian nerve cords. Nature 553(7686):45–50
Martindale MQ (2005) The evolution of metazoan axial properties. Nat Rev Genet 6(12):917–927
Mayr E (1982) The growth of biological thought: Diversity, evolution, and inheritance. Harvard University Press
Nahmad M, Glass L, Abouheif E (2008) The dynamics of developmental system drift in the gene network underlying wing polyphenism in ants: a mathematical model. Evol Dev 10(3):360–374
Nejad Kourki A (2021) Beyond Congruence: Evidential Integration and Inferring the Best Evolutionary Scenario. Preprints. doi:https://doi.org/10.20944/preprints202105.0767.v1
Niehrs C (2010) On growth and form: a Cartesian coordinate system of Wnt and BMP signaling specifies bilaterian body axes. Development 137(6):845–857
Nielsen C (2012) Animal evolution: interrelationships of the living phyla. Oxford University Press on Demand
Nielsen C (2019) Early animal evolution: a morphologist’s view. Royal Soc open Sci 6(7):190638
Nielsen C, Brunet T, Arendt D (2018) Evolution of the bilaterian mouth and anus. Nat Ecol Evol 2(9):1358–1376
Nielsen C, Martinez P (2003) Patterns of gene expression: homology or homocracy? Dev Genes Evol 213(3):149–154
Novick A (2018) The fine structure of ‘homology’. Biology & Philosophy 33(1–2):6
Panganiban G, Irvine SM, Lowe C, Roehl H, Corley LS, Sherbon B, Walker M (1997) The origin and evolution of animal appendages. Proceedings of the National Academy of Sciences, 94(10), 5162–5166
Patterson C (1982) Morphological characters and homology. Problems of phylogenetic reconstruction, 21–74
Patterson C (1988) Homology in classical and molecular biology. Mol Biol Evol 5(6):603–625
Peterson KJ, Cameron RA, Davidson EH (2000) Bilaterian origins: significance of new experimental observations. Dev Biol 219(1):1–17
Pisani D, Pett W, Dohrmann M, Feuda R, Rota-Stabelli O, Philippe H, Wörheide G (2015) Genomic data do not support comb jellies as the sister group to all other animals. Proceedings of the National Academy of Sciences, 112(50), 15402–15407
Prum RO, Brush AH (2002) The evolutionary origin and diversification of feathers. Q Rev Biol 77(3):261–295
Ramsey G, Siebels Peterson A (2012) Sameness in biology. Philos Sci 79(2):255–275
Remane A (1956) Die Grundlagen des natürlichen Systems, der vergleichenden Anatomie und der Phylogenetik: theoretische Morphologie und Systematik
Rieppel O (1994) Homology, topology, and typology: the history of modern debates. Homology. Elsevier, pp 63–100
Sanetra M, Begemann G, Becker M-B, Meyer A (2005) Conservation and co-option in developmental programmes: the importance of homology relationships. Front Zool 2(1):1–17
Schäfer K (2003) Origination of organismal form: beyond the gene in developmental and evolutionary biology, vol 2. MIT Press
Shubin N, Tabin C, Carroll S (1997) Fossils, genes and the evolution of animal limbs. Nature 388(6643):639–648
Shubin N, Tabin C, Carroll S (2009) Deep homology and the origins of evolutionary novelty. Nature 457(7231):818–823
Stadler PF, Stadler BM (2006) Genotype-phenotype maps. Biol Theory 1(3):268–279
Tarazona OA, Lopez DH, Slota LA, Cohn MJ (2019) Evolution of limb development in cephalopod mollusks. Elife 8:e43828
True JR, Haag ES (2001) Developmental system drift and flexibility in evolutionary trajectories. Evol Dev 3(2):109–119
Van Valen LM (1982) Homology and causes. J Morphol 173(3):305–312
Wagner GP (2007) The developmental genetics of homology. Nat Rev Genet 8(6):473–479
Wagner GP (2014) Homology, genes, and evolutionary innovation. princeton university press
Wijesena N, Simmons DK, Martindale MQ (2017) Antagonistic BMP–cWNT signaling in the cnidarian Nematostella vectensis reveals insight into the evolution of mesoderm. Proceedings of the National Academy of Sciences, 114(28), E5608-E5615
Williams DM, Ebach MC (2020) Cladistics. Cambridge University Press
Wray GA (2007) Evolutionary dissociations between homologous genes and homologous structures. Paper presented at the Novartis Foundation Symposium 222-Homology: Homology: Novartis Foundation Symposium 222
Wray GA, Abouheif E (1998) When is homology not homology? Curr Opin Genet Dev 8(6):675–680
Zhao Y, Vinther J, Parry LA, Wei F, Green E, Pisani D, Cong P (2019) Cambrian sessile, suspension feeding stem-group ctenophores and evolution of the comb jelly body plan. Curr Biol 29(7):1112–1125e1112
Acknowledgements
I would like to thank my former supervisor Professor Philip C. J. Donoghue (Bristol Palaeobiology) for instilling some of the early ideas in this paper and giving the initial motivation, as well as Professor Günter P. Wagner (Yale), Dr James DiFrisco (Leuven), Professor Graham Budd (Uppsala), Dr Jordi Paps Montserrat (Bristol), Dr Geoffrey Keeling (Stanford), and two anonymous reviewers for invaluable biological and philosophical comments on this paper.
Funding
This research was undertaken as part of my PhD project at the University of Bristol, partially funded by the university’s fee waiver scholarship for international students.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest/Competing interests
N/A.
Ethics approval
N/A.
Consent to participate
N/A.
Consent for publication
N/A.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nejad Kourki, A. The integrative approach to inferring homology: morphology and development combined. Biol Philos 37, 26 (2022). https://doi.org/10.1007/s10539-022-09846-1
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10539-022-09846-1