Abstract
Bisphenol A (BPA) is an endocrine disrupting chemical. Its extensive use has led to the wide occurrence of BPA in various environmental ecosystems, at levels that may cause negative effects to the ecosystem and public health. Although there are many bacteria able to BPA utilization, only a few of them have a strong capacity for its biodegradation. Therefore, it is important to search for new bacteria strains, investigate their BPA biodegradation ability and potential effect of pH and other organic compounds on the process. These tasks have become the object of the present study. The results of our research show that for the newly isolated strains Acinetobacter sp. K1MN and Pseudomonas sp. BG12 after 15 days, with an initial BPA concentration of 100 mg L− 1, the highest BPA removal was achieved at pH 8, while sodium glutamate as a biostimulant best accelerated BPA degradation. Kinetic data for BPA biodegradation by both strains best fitted the Monod model. The specific degradation rate and the half saturation constant were estimated respectively as 8.75 mg L− 1 day− 1 and 111.27 mg L− 1 for Acinetobacter sp. K1MN, and 8.6 mg L− 1 day− 1 and 135.79 mg L− 1 for Pseudomonas sp. BG12. The half-maximal effective concentration (EC50) of BPA for Acinetobacter sp. K1MN was 120 mg L− 1 and for Pseudomonas sp. BG12 it was 123 mg L− 1. The toxicity bioassay (Microtox test) showed that elimination of BPA by both strains is accompanied by reduction of its toxic effect. The ability of tested strains to degrade BPA combined with their high resistance to this xenobiotic indicates that Acinetobacter sp. K1MN and Pseudomonas sp. BG12 are potential tools for BPA removal during wastewater treatment plant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bisphenol A (BPA; 2,2-bis(4-hydroxyphenyl)propane) is an endocrine-disrupting chemical (EDC) capable of interfering with the function of sex hormones, insulin, leptin, and thyroxin (García-Espiñeira et al. 2018). It can also induce immunotoxic, mutagenic, genotoxic, hepatotoxic, teratogenic, neurotoxic and carcinogenic effects, even at nanomolar level (Pfeifer et al. 2015).
Despite BPA’s negative impact on the human body, it is one of the most commonly produced and used compounds worldwide with annual production expected to reach 10.6 million metric tons in 2022. Its annual growth rate between 2016 and 2022 is approximately 4.8% (Industry Experts, 2016). Because of the wide usage of polycarbonate plastics and epoxy resins in industry and households, BPA is a prevalent contaminant in the environment and its concentration, especially in the aquatic environment, is constantly increasing (Cleveland et al. 2014; Bilal et al. 2019; Grelska and Noszczyńska 2020). It enters these ecosystems mainly through the effluents of wastewater treatment plants (WWTPs), where by lack of efficient systems of its removal, BPA may contaminate drinking water sources downstream (Zielinska et al. 2019).
Taking into account that BPA possesses an ecological risk, there is an urgent necessity to eliminate it from the environment. One of the ways to remove BPA from ecosystems is its microbial degradation. Therefore, searching for efficient BPA degraders and detailed studies on microbial utilization of BPA are vital to engineer methods that enable its effective elimination from different environments compartments. A large number of bacteria capable of BPA degradation have been isolated from different environments such as rivers, seawater, wastewaters, leachates, sludges, soil, desert soil and the rhizosphere of plants (Kang and Kondo 2002; Sasaki et al. 2005a, b; Toyama et al. 2009; Fischer et al. 2010; Zühlke et al. 2016; Kamaraj et al. 2018; Suyamud et al. 2018; Louati et al. 2019). In some of these bacteria, a few enzymes active in BPA utilization have been identified. A cytochrome P450 monooxygenase was reported to catalyze the transformation of BPA to BPA-M and the BPA ipso substitution in Sphingomonas sp. AO1 and Sphingomonas sp. TTNP3, respectively (Sasaki et al. 2005a, b; Kolvenbach et al. 2014). An ammonia monooxygenase in Nitrosomonas europaea and an extracellular laccase in Pseudomonas sp. LBC1 were also identified to be involved in BPA utilization (Kolvenbach et al. 2007; Roh et al. 2009; Telke et al. 2009). Also proteins engaged in protocatechuate transformation are probably involved in the BPA degradation pathway in Sphingobium sp. BiD32. Moreover, a p-hydroxybenzoate hydroxylase, which likely takes part in metabolism and degradation of xenobiotics, was also linked with BPA degradation by the strain BiD32 (Zhou et al. 2015).
Most of these abovementioned bacterial isolates degraded about 70% of BPA in medium containing up to 1 mg L− 1 of the compound (Kang and Kondo 2002; Sasaki et al. 2005a, b; Toyama et al. 2009; Fischer et al. 2010; Zühlke et al. 2016; Kamaraj et al. 2018; Suyamud et al. 2018; Louati et al. 2019). At a relatively high concentration of BPA, the efficiency of bacterial degradation decreased (Fischer et al. 2010; Kamaraj et al. 2014; Heidari et al. 2017).
Taking this into account and the constantly increasing amount of BPA in the aquatic environments, it is necessary to search for new pure bacterial strains that have potential to degrade BPA in high concentration what may lead to the development of successful biodegradation strategy of this compound which could be applied in WWTPs. However, in wastewater, apart from xenobiotics, other organic compounds are present. These compounds might served as additional growth substrates for bacteria and might affect their enzyme stability hence changing bacteria’s potential to degrade xenobiotics. Another factor which determine the enzyme stability is pH. Therefore, the main novelty of the present study was to determine how selected additional growth substrates and pH influence on BPA degradation rate by newly isolated bacteria utilizing BPA at concentration of 100 mg L− 1. We also estimated the kinetic parameters of the degradation process, determined the inhibitory effect of BPA on bacterial growth, and analyse BPA toxicity before and after degradation by the isolated strains.
Materials and methods
Reagents and media
Bisphenol A and ethyl acetate (HPLC grade) were obtained from Merck (Darmstadt, Germany). HPLC grade acetonitrile and ethanol were purchased from S. Witko - JT Baker (Lodz, Poland). Water used as a HPLC solvent was purified with a Direct-Q® Water Purification System (Merck). The standard stock solutions of BPA (7 g L− 1 or 5 mg 5 mL− 1) were prepared in 70% ethanol and stored at 4 °C up to three months. Final concentration of BPA and ethanol in medium were 100 mg L− 1 and 1% (v/v), respectively. Basal Salt Medium (BSM) (Badiefar et al. 2015) was used for isolation and purification of bacterial species and the degradation study. For bacteria isolation, BSM was supplemented with nystatin (4 g L− 1) and actidione (4 g L− 1) to inhibit the growth of fungi.
Sampling and isolation of bisphenol A degrading bacteria
Samples used for isolation of BPA-degrading bacteria were collected from WWTP Klimzowiec, Katowice (1000 ml of activated sludge), a landfill in Tychy (1000 mL of leachate), Petrochemia-Blachownia SA, Kędzierzyn Koźle (10 g of soil) and Kalina pond, Świętochłowice (1000 ml of water with sediment).
The activated sludge, leachate and water with sediment were centrifuged (4700 rpm, 20 min, 4 °C). Supernatants were discarded and pellets were re-suspended in 20 mL of sterile Millipore Water. 10 mL of these suspensions and 10 g of the collected soil were added to separate flasks containing 90 mL of BSM supplemented with BPA at a final concentration of 20 mg L− 1 and incubated at 28 °C with rotary shaking (120 rpm). After 7 days, 10 mL of acclimated consortiums were used as inocula to start fresh batches with gradually increasing concentrations of BPA (40–100 mg L− 1). The morphologically distinct bacterial strains were isolated and screened for strains with the highest BPA degradation abilities. Briefly, the selected strains were cultivated in 100 mL of BSM enriched with 100 mg L− 1 BPA, at 28 °C and 120 rpm for six days. Then, 1 mL of each culture was collected and BPA concentration was measured using high-performance liquid chromatography (HPLC). The two most effective BPA degraders named K1MN and BG12 were selected for further experiments. The concept of these experiments is presented in Fig. 1.
Identification of selected strains
The two selected strains K1MN isolated from Kalina pond and BG12 isolated from soil were morphologically and phenotypically characterized (API 21 system, BioMerieux, Lyon, France).
Genomic DNA was extracted from K1MN and BG12 using a DNA extraction kit (Bacterial & Yeast Genomic DNA Purification Kit, EURx) according to the manufacturer’s instructions. The 16S rRNA gene sequence was amplified with primers 8F and 1492R targeting a fragment size of 1484 bp (Pacwa-Płociniczak et al. 2014). The PCR reaction contained: 1 µL of the DNA template, 0.125 µL DreamTaq DNA polymerase (5 U/µL) (Thermo Fisher Scientific), 2.5 µL 10 × DreamTaq Buffer (Thermo Fisher Scientific), 1 µL dNTP Mix (10 mM; Thermo Fisher Scientific, Invitrogen™), 1 µL 16S Forward Primer (0.1 µg/mL), 1 µL 16S Reverse Primer (0.1 µg/mL), 25 µL reaction. PCR amplification was performed at 95 °C for 5 min, 3 cycles at 94 °C for 45 s, 57 °C for 30 s, 72 °C for 120 s; 3 cycles at 95 °C for 45 s, 56 °C for 30 s, 72 °C for 120 s; 3 cycles at 95 °C for 45 s, 56 °C for 30 s, 72 °C for 120 s; 31 cycles at 95 °C for 45 s, 53 °C for 30 s, 72 °C for 120 s; and a final elongation cycle at 72 °C for 5 min in a C1000 Touch™Thermal Cycler (BioRad). Gene sequencing was performed by an external company (Genomed, Poland). The obtained sequences (1417 bp for K1MN and 1411 bp for BG12) were compared with EZBioCloud database. The phylogenetic analysis was done based on the longest common fragment of the 16S rRNA gene sequences selected from ClustalW alignment of K1MN and BG12 strains and closest type strains of other Acinetobacter and Pseudomonas species, respectively which were obtained from GenBank (Larkin et al. 2007; Furmanczyk et al. 2018). Phylogenetic analysis and evolutionary distance calculations were determined using the maximum-likelihood method based on 1000 bootstrap resampling and a Tamura 3-parameter model assuming that a certain fraction of sites are evolutionarily invariable (+ I) conducted using Mega X software (Furmanczyk et al. 2018; Kumar et al. 2018). The determination of the similarity of 16S rRNA sequences between tested and type strains was done using the Average Nucleotide Identity (ANI) calculator (https://www.ezbiocloud.net/tools/ani) (Płociniczak et al. 2019).
Effects of pH and additional growth substrate on BPA-degrading activity of the strains
Degradation experiments were performed in Erlenmeyer flasks containing 250 mL of BSM and BPA (100 mg L− 1) at pH 7.2. The flasks were supplemented with glucose, sucrose, monosodium glutamate (1 g L− 1) or phenol (5%) to evaluate the effect of the additional carbon sources as biostimulants on BPA degradation. Each created set was made in triplicate and inoculated either with 100 µL of the K1MN or BG12 cells being at logarithmic growth phase (log-phase cells) (OD600 = 0.3). As a control non-inoculated media were used. The biodegradation experiments were maintained for 15 days at 28 °C, pH 7.2 with rotary shaking, 120 rpm. Samples were collected periodically every 5 days to determine the growth of studied strains (OD600) and evaluate BPA concentration.
In order to study the effect of pH value on BPA degradation, 250 mL of BSM with BPA (100 mg L− 1) were adjusted to pH in the range 3–8. All media at given pH were made in triplicate and inoculated with log-phase cells and the experiment was carried out according to the procedure described above.
Half-maximal effective concentration (EC50)
To elucidate the inhibitory effect of BPA on the growth of tested strains, log-phase cells were adjusted to the optical density 0.05 and 100 µl of the suspension was transferred to the nutrient broth supplemented with BPA in the concentration range of 20–320 mg L− 1. Each set was prepared in triplicate. After 24 h cultivation with rotary shaking, 120 rpm, at 28 °C, the OD600 of the cultures was measured. The EC50 value was calculated using five parameter logistic regression with SigmaPlot 14.0 software. The equation is presented below
where
\(min\) is the bottom of the curve; \(max\) is the top of the curve; \(Hillslope\) characterizes the slope of the curve at its midpoint; \(s\) is the asymmetry parameter; \(x\) is the BPA concentration, \(y\) is the optical density of the bacterial culture.
Determination of kinetic parameters of bisphenol A degradation
For analysis of the degradation kinetics of BPA, 0.01 g of log-phase cells of K1MN and BG12 strains were separately inoculated in a series of 300 mL Erlenmeyer flasks containing 150 mL of BSM supplemented with BPA at initial concentrations of 1, 10, 30, 60, 100, 120 or 140 mg L− 1. This range of concentrations was selected based on the obtained results in experiment described in Sect. 2.5. Each set of flasks was prepared in triplicate. Flasks were incubated for 30 days at 28 °C with shaking at 100 rpm. Every 24 h, growth of strains was monitored by OD600 measurement and the concentration of residual BPA in the medium was determined. For studying biodegradation of BPA, the Monod model was used, which is presented by the following equation (Eq. 1):
where S is the substrate concentration (mg L− 1), Vmax is the maximum specific BPA degradation rate (mg L− 1 day− 1), Ks is the half saturation constant (mg L− 1).
Kinetic constants were estimated using SigmaPlot 12.0 software.
Toxicity bioassay
The acute toxicity of the initial and residual BPA concentration after 30-days degradation by K1MN and BG12 strains was evaluated by the Microtox test. The initial concentration of BPA (100 mg L− 1) was selected on the basis of EC50 and Ks values obtained for both strains. Each setup was done in triplicate. The assay was performed in accordance with the manufacturer’s procedures using freeze-dried Vibrio fischeri NRRL B-11,177 and Microtox Model 500 Analyser (Modern Water Inc., UK). The inhibition of the luminescence of NRRL B-11,177 was compared to the control sample (bacteria not treated with BPA) after 5 and 15 min of exposure. The standard protocol 81.9% Basic Test was done during which all samples were diluted with 2% NaCl at 81.9% of the initial sample concentration. The toxicity units (TU = 1/EC50 − 1 × 100) were calculated using the MicrotoxOmni (Microbics Corp, 1992) program (Le et al. 2017).
Analytical methods
For the determination of BPA concentration, 1 mL of cultures was collected and centrifuged (14,000 rpm, 10 min). 1 mL of ethyl acetate was added to the resultant supernatants and the mixtures were vortexed at 3000 rpm for 60 s followed by centrifugation (1000 rpm, 60 s). Organic layers were transferred to Pyrex glass and dried under a stream of N2. 1 mL of 70% ethanol was added to the pellets and samples were vortexed at 3000 rpm for 60 s. Solutions were filtered through 0.2 µm RC membrane filter (Hahnemuehle, Germany) and used for detection and measurement of BPA content using a Shimadzu HPLC (Kyoto, Japan) with quaternary pumps (model LC-20AD), connected to a PDA detector (Shimadzu, model SPD-M20A) interfaced with the LabSolutions software. Separation was achieved using the Phenomenex Synergi 4 µm Hydro-RP (150 × 4.6 mm) column, protected by an AQ C18 guard column (Phenomenex, Torrance CA, USA). The samples were eluted with a linear gradient of acetonitrile–water (90−10%) with a flow rate of 1 mL min− 1 for 35 min. The column temperature was maintained at 30 °C. The injection volume was 15 µL. All experiments were carried out in triplicate and results are expressed as an average value.
The calibration curve was obtained from a linear regression program by concentrations versus detector responses using concentration levels for eight standards. These working solutions were prepared from stock solution of 1 g L− 1 at concentrations of 0.5, 10, 50, 100, 200, 400, 600 and 1000 mg L− 1. The correlation coefficient of peak height to concentration was > 0.998.
The percentage BPA removal efficiency (RE) was calculated using the equation:
where Co and Cf are the initial and final concentrations of BPA (mg L− 1) in BSM, respectively.
Statistical analysis
In order to check the normality of the data for all of the results obtained in Sects. 2.4 and 2.7, the Shapiro-Wilk tests was used. A one-way or two-way ANOVA analysis (p < 0.05) followed by a Fisher’s least significant difference (LSD) test was performed to conduct the statistical significance. Statistical analysis was done using STATISTICA 13.1 PL software (StatSoft, Tulsa, USA).
Results and discussion
Isolation and identification of selected strains
Forty bacterial strains able to grow in the presence of 100 mg L−1 BPA as a sole carbon source were isolated after 5 weeks of incubation. The BPA degrading activities of these strains were determined as a loss of BPA (100 mg L−1) amount in BSM after 6 days by HPLC analysis. All isolates were able to grow in the medium and degraded BPA with different efficiency (data not shown). For two strains named K1MN and BG12 the RE was higher than to the other strains and was 20 ± 3% and BG12 36 ± 2%, respectively. These strains were isolated from Kalina pond (K1MN) and soil from Petrochemia-Blachownia SA area (BG12).
The strains were characterized by morphological and biochemical assays (Table 1 in supplementary data). The bacteria were rod-shaped and Gram-negative. Strain K1MN similar to Acinetobacter johnsonii was positive for citrate utilization and negative for gelatin hydrolysis as well as assimilation of glucose and arabinose (Kozińska et al. 2014; Juni 2015). Strain BG12 assimilated glucose and was negative for nitrate reduction, analogously to Pseudomonas protegens strain CHA0 (Ramette et al. 2011).
The phylogenetic analysis based on the ClustalW alignment of the 16S rRNA gene sequences of strain K1MN (1283 bp) and the 17 closest type strains of other Acinetobacter species placed it in the A. johnsonii subgroup (Fig. 2).
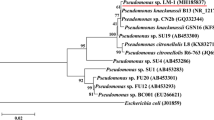
The analysis of the BG12 sequence (1333 bp) and its 20 closest type Pseudomonas strains revealed that it is clustered in the P. protegens subgroup (Fig. 3). To verify the phylogenetic relationship of isolated strains, the ANI was calculated among them and related species. These results confirmed that the closest related species of strain K1MN is A. johnsonii ATCC 17,909 with similarity of 99.10%. The next closest related species are: A. bouvetii DSM 14,964 with 98.20%, A. lwoffi DSM 2403 with 96.80%, A. kyonggiensis KSL5401-037 with 96.65%, and A. albensis ANC 4874 with 95.70%. In the case of the BG12 strain, the sequence of the 16S rRNA gene showed 99.90% similarity with P. protegens CHA0, 99.0% similarity with P. sesame SI-P133 and 98.40% similarity with P. saponiphila DSM 9751.
Based on morphologic, phenotypic, ANI and partial 16S rRNA analyses, isolates were identified as Acinetobacter sp. strain K1MN and Pseudomonas sp. strain BG12.
Phylogeny of type strains closely related to K1MN strain based on 16S rRNA gene sequence. All positions containing gaps or missing data were eliminated, which resulted in a 1283 bp sequence in the final dataset. Bootstrap values are represented at the branching points. The bar represents 0.05 substitutions per site. Accession numbers of sequences used in this analysis are in parentheses
Phylogeny of type strains closely related to BG12 strain based on 16S rRNA gene sequence. All positions containing gaps or missing data were eliminated, which resulted in a 1333 bp sequence in the final dataset. Bootstrap values are represented at the branching points. The bar represents 0.020 substitutions per site. Accession numbers of sequences used in this analysis are in parentheses
Effects of pH and additional growth substrate on BPA-degrading activity of the strains
As it has been mentioned before, Acinetobacter sp. K1MN and Pseudomonas sp. BG12 were able to degrade 20 ± 3% and 36±%2 BPA at an initial concentration of 100 mg L− 1, respectively within 15 days. Since pH values strongly influence the activity of most enzymes, their impact on BPA degradation capacity of the strains was studied. Moreover, in the wastewaters besides from xenobiotics other various organic compounds are present. On the one hand, some of them might blocking the active sites of degradation enzymes thus inhibiting degradation processes (Górny et al. 2019). On the other hand, some organic compounds contribute to the growth of bacterial cells and the faster degradation of xenobiotics by being as an additional carbon source for bacteria and/or by producing specific monooxygenase enzymes by the bacteria in the case of the structural analogue of the degraded pollutant (Domaradzka et al. 2015; Górny et al. 2019). Therefore, it was interesting to verify whether pH and additional growth substrate such as phenol, glucose, saccharose and sodium glutamate can affect the degradation capacity of the tested strains.
Figure 4 shows the degradation trends of BPA in the pH range from 3 to 8. For both strains, complete BPA degradation was not achieved at any of the pH levels. At pH from 3 to 6 the RE of BPA was 4–11% and 4–16% for Acinetobacter sp. K1MN and Pseudomonas sp. BG12, respectively. It was associated with a low survival rate of the strains in an acidic environment, resulting in reduced activity of degrading enzymes or even their lack (Li et al. 2012). The highest degradation values of 60% for pH 8 and 35% for pH 7 were obtained for Pseudomonas sp. BG12. In contrast, under these pH values a slight BPA reduction (21%) was observed for Acinetobacter sp. K1MN. Under weak alkaline conditions, the growth of bacteria was not inhibited and BPA was more soluble due to its hydrolysis. Moreover, at these pH values, the surface of the bacterial cells may have been negatively charged, leading to changes in electrostatic interaction between BPA and the biomass surface (Wolski et al. 2006). Such conditions result in higher BPA degradation efficiency (An et al. 2011)
Degradation efficiency (Acinetobacter sp. K1MN – a, Pseudomonas sp. BG12 – c) and strains’ growth monitored as optical density at 600 (Acinetobacter sp. K1MN – b, Pseudomonas sp. BG12 – d) in BSM medium with BPA (100 mg L− 1). The data points represent the average of three independent experiments ± standard deviation. The same letter(s) above the bars indicate no statistical significance (MANOVA followed by Fisher’s LSD test) related to the effects of BPA degradation and optical density of cultures at p < 0.05
These findings differ from those of Li et al. (2012), who demonstrated that Bacillus sp. GZB degraded BPA in 96 h in Luria-Bertani medium at pH 6.5, 7.0, 8.0, 8.5 and 9.0 with efficiencies of 87.5%, 100%, 100%, 97.7% and 81.6%, respectively. Such high RE resulted from the low initial BPA concentration (10 mg L− 1) and use of a rich medium. At the same initial BPA concentration but in mineral medium at pH 7, strains Pseudomonas sp. K-8, K-6 and KU-3 degraded BPA in 12 days with RE 81%, 78% and 74%, respectively (Kamaraj et al. 2014). By contrast, Heidari et al. (2017) observed that Ralstonia eutropha was not able to complete BPA removal even at low concentrations ranging from 1 to 20 mg L− 1 in mineral medium at pH 7. After 12 days, the BPA RE was 15–56%. Overall, these results showed, that the most suitable BPA concentration for its effective degradation is up to 10 mg L− 1 in rich medium at pH of 7 or 8. This is in agreement with previous studies showing that medium composition may play an important role in complete BPA removal (Badiefar et al. 2015).
Degradation efficiency (Acinetobacter sp. K1MN - a, Pseudomonas sp. BG12 - c) and strains growth monitored as optical density at 600 (Acinetobacter sp. K1MN - b, Pseudomonas sp. BG12 - d) in BSM medium with BPA (100 mg L− 1) and in the presence of different, additional substrates. Controls contained no additional source of carbon. The data points represent the average of three independent experiments ± standard deviation. The same letter(s) above the bars indicate no statistical significance (MANOVA followed by Fisher’s LSD test) related to the effects of BPA degradation and optical density of cultures at p < 0.05
In Fig. 5 the degradation efficiency of BPA by the two tested strains in conditions with additional carbon source was shown. Due to the chemical structure similarity of phenol and BPA, it was likely that phenol would induce synthesis of the enzymes engaged in aromatic ring fission (Heidari et al. 2017). Glucose, saccharose and sodium glutamate proved to accelerate contaminant removals (Kamaraj et al. 2014; Zhao et al. 2014; Marchlewicz et al. 2017; Górny et al. 2019). However, in our study glucose and phenol caused inhibition of both BPA degradation and bacterial growth of Acinetobacter K1MN (Fig. 5a, b). These findings are not consistent with results reported by others who have shown that phenol stimulated growth of Cupriavidus basilensis JF1 but not BPA degradation, while BPA removal by Bacillus sp. GZB was enhanced by adding glucose (Fisher et al. 2010; Xiong et al. 2017). On the other hand, BPA RE decreased when R. eutropha was grown in the presence of phenol, while glucose did not affect the degradation rate of Pseudomonas sp. strains KU1 and KU2 as well as Bacillus sp. KU2 (Kamaraj et al. 2014; Babatabar et al. 2019). Similarly, saccharose did not have an influence on BPA degradation by Acinetobacter K1MN.
The highest BPA RE was observed in the presence of sodium glutamate and reached the value of 31 ± 2.12% and 52 ± 4.58% for Acinetobacter sp. K1MN and Pseudomonas sp. BG12, respectively (Fig. 4). The addition of the substrate had a positive effect on BPA degradation efficiency in comparison to monosubstrate culture, where only 21 ± 4% (Acinetobacter sp. K1MN) and 35 ± 4.24% (Pseudomonas sp. BG12) of BPA was degraded after 15 days. We assume that the presence of sodium glutamate in the culture medium increased the tolerance of the strains to high BPA concentrations more than other tested substrates by providing a good source of readily metabolizable carbon and nitrogen to support cell growth. The findings are directly in line with previous findings. Kamaraj et al. (2014) showed that in the presence of sodium glutamate degradation of phenol by Pseudomonas sp. JN-6 increased by 10% while BPA RE of Pseudomonas sp. K-8 increased to 90%. The addition of glucose and sucrose caused a statistically significant decrease of BPA RE in the case of Pseudomonas sp. K1MN, while phenol completely inhibited BPA degradation by this strain (Fig. 5c, d). The negative effects of phenol on the BPA biodegradation process as well as biomass growth of both strains probably result from its toxicity and not adapting the tested strains to grow in its presence. From the presented results, it is clear that both tested strains were able to survive and utilize BPA at the concentration of 100 mg L− 1. Therefore, further studies (Fig. 6) were undertaken to better characterize the ability of both strains to degrade BPA.
Half maximal effective concentration (EC50)
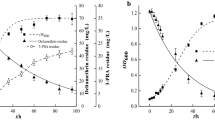
In order to analyse if ability of K1MN and BG12 strains to degrade BPA at the concentration of 100 mg L− 1 is correlated with the higher resistance of these strains to the compound, EC50 values were calculated based on growth inhibition curves by various concentrations of BPA. Effective concentration in this case means the BPA concentration causing 50% growth inhibition of particular strain. Both used strains tolerate relatively high BPA concentrations. EC50 values of Acinetobacter sp. K1MN and Pseudomonas sp. BG12 were 120 mg L− 1 and 123 mg L− 1 BPA, respectively (Fig. 7). The concentration of the compound that completely inhibited growth of Acinetobacter sp. was 270 mg L− 1. Pseudomonas sp. BG12 tolerates higher concentrations of BPA and its growth was totally inhibited by 300 mg L− 1 BPA in nutrient broth medium. The results show that tested strains tolerate high BPA concentrations. It is probably caused by adaptation of the previous strain to the presence of BPA (100 mg L− 1) in medium and its capacity for BPA degradation. Probably as with phenol, an increase in BPA leads changes in the cell membrane and protect the cell from its toxic effects (Murínová and Dercová 2014). To compare, the EC50 value for Cupriavidus basilensis JF1 was established as 0.12 mM, equivalent to 27.39 mg L− 1 (Fischer et al. 2010). Reports regarding higher water organisms indicate stronger toxicity of BPA. For example, EC50 after 24 h of Daphnia magna exposure to BPA was estimated as 8.9 mg L− 1 (Tišler et al. 2016) and for the microalga Cyclotella caspia, 96 h EC50 was determined as approximately 8 mg L− 1 BPA (Li et al. 2008).
Determination of kinetic parameters of BPA degradation
For calculating the specific degradation rate (SDR) for Acinetobacter sp. K1MN and Pseudomonas sp. BG12, BPA concentration in a series of separate flask containing different BPA concentrations from 1 to 140 mg L− 1 with the initial biomass concentration of 0.01 g L− 1 was monitored throughout the thirty-day experiment (Babatabar et al. 2019). SDR was determined by dividing the degradation rate by the initial biomass for each initial BPA concentration. Figure 8 shows the obtained SDRs values. Different kinetic models were used to fit the experimental data (Robinson and Tiedje 1983; Okpokwasili and Nweke 2006). Among them, the Monod model gave the best fit with R2 = 0.97 for Acinetobacter sp. K1MN and R2 = 0.84 for Pseudomonas sp. BG12. Therefore, this model was used, however, the fit is not clear hence the Vmax and Ks values (Table 1) might be imprecise.
The Monod equation for BPA biodegradation by Acinetobacter sp. K1MN can be represented by Eq. 2, and by Pseudomonas sp. BG12 by Eq. 3.
X represents the biomass concentration (g L− 1).
The Vmax values obtained for both strains are comparable to that of Ralstonia eutropha adapted for the growth of 20 mg BPA where Vmax = 7.4 mg L− 1, but significantly different from the results of Vmax = 0.46 mg L− 1 obtained for Sphingomonas paucimobilis FJ-4 (Fujiwara et al. 2016; Heidari et al. 2017). Thus, it is concluded that both Acinetobacter sp. K1MN and Pseudomonas sp. BG12 show great ability in BPA degradation.
Toxicity of medium containing BPA
Even though BPA has been detected at concentrations ranging from nanograms per liter (ng L− 1) to micrograms per liter (mg L− 1) in drinking water, the long-term continuous exposure of a living organism to this EDC cannot be ignored considering its harmful impact (Sarma and Lee 2018). Therefore, it was interesting to estimate the toxicity of medium containing BPA (100 mg L− 1) and this medium inoculated with strains K1MN and BG12 after a 30-day cultivation period. For this purpose a Microtox bioassay, a sensitive system standardized for water and effluent samples, was chosen for toxicity evaluation (Lei and Aoyama 2010). Toxicity of each sample was determined as toxicity units (TU). An increase in TU value corresponds to an increase in toxicity (Biedroń et al. 2016). The medium with BPA after 5 and 15 min contact with V. fischeri showed high acute toxicity, being TU50 = 125.35 ± 1.7 and TU50 = 124.75 ± 1.2, respectively (Fig. 2 in supplementary data). After 30 days of degradation by Acinetobacter sp. K1MN, toxicity of medium with BPA was reduced to TU50 = 52.96 ± 10.99 and TU50 = 60.31 ± 14.13, respectively after 5 and 15 min of exposure. In contrast, metabolic activity of Pseudomonas sp. BG12 led to reduction of this medium toxicity to TU50 = 8.83 ± 2.05 after 5 min and TU50 = 10.34 ± 4.13 after 15 min of exposure. This result has been supported by HPLC analysis which revealed that Pseudomonas sp. BG12 removed 15% more of the initial BPA concentration than Acinetobacter sp. K1MN (data not shown). The differences in media toxicity between the two strains are probably due to the higher RE of BPA degradation by BG12 than by K1MN. This result is relevant as it demonstrates that the strains, especially Pseudomonas sp. BG12, are able to degrade BPA, which is correlated with a significant decrease in toxicity of the medium. It corresponds well with the research of Ike et al. (2002), who revealed that biodegradation can remarkably reduce the toxic effects of BPA. However, the main limitation of the cited study was the use of commercially available end products of one of the BPA degradation pathways, while it can be biological decomposed during various mechanisms (Noszczyńska and Piotrowska-Seget 2018). Conversely, others have shown that even when organisms degrade BPA with high RE, intermediates formed during degradation are often more toxic than the parent compound (Mtibaà et al. 2018). Similarly, chemical technologies such as photocatalytic oxidation did not reduce BPA toxicity but even increased it in comparison with untreated BPA solution (Plahuta et al. 2014).
Conclusion
BPA is one of the most abundant pollutants in the aquatic environment and can affect surface and groundwater systems. Due to its negative impact on living organisms, it is very important to select microorganisms with the ability to decompose BPA. Acinetobacter sp. K1MN and Pseudomonas sp. BG12 seem to fulfil this expectation. Both strains showed the capacity for BPA removal. Biodegradation of this EDC was enhanced in the alkaline conditions and in the presence of monosodium glutamate. Simultaneously, other additional substrates had no positive effect on degradation ability of the strains. Promising application of the examined strains in the treatment of BPA contaminated water is related to their tolerance of high BPA concentrations and significant reduction of its toxicity. Moreover, the data from our investigation provide insight into the influence of environmental factors on BPA elimination from ecosystems.
References
An T, Zu L, Li G et al (2011) One-step process for debromination and aerobic mineralization of tetrabromobisphenol-A by a novel Ochrobactrum sp. T isolated from an e-waste recycling site. Bioresour Technol 102:9148–9154. https://doi.org/10.1016/j.biortech.2011.06.080
Babatabar S, Zamir SM, Shojaosadati SA et al (2019) Cometabolic degradation of bisphenol A by pure culture of Ralstonia eutropha and metabolic pathway analysis. J Biosci Bioeng 127:732–737. https://doi.org/10.1016/j.jbiosc.2018.12.001
Badiefar L, Yakhchali B, Rodriguez-Couto S et al (2015) Biodegradation of bisphenol A by the newly-isolated Enterobacter gergoviae strain BYK-7 enhanced using genetic manipulation. RSC Adv 5:29563–29572. https://doi.org/10.1039/C5RA01818H
Biedroń I, Chojniak J, Sobolczyk J, Jalowiecki Ł (2016) The use of the microtox system to assess the potential release of toxic substances from the sorption materials used in drinkig water treatment. Water Resour For Mar Ocean Ecosyst Conf Proceedings 1:773–779. https://doi.org/10.5593/SGEM2016/B31/S12.101
Bilal M, Iqbal HMN, Barceló D (2019) Mitigation of bisphenol A using an array of laccase-based robust bio-catalytic cues: a review. Sci Total Environ 689:160–177. https://doi.org/10.1016/j.scitotenv.2019.06.403
Cleveland V, Bingham JP, Kan E (2014) Heterogeneous Fenton degradation of bisphenol A by carbon nanotube-supported Fe3O4. Sep Purif Technol 133:388–395. https://doi.org/10.1016/j.seppur.2014.06.061
Domaradzka D, Guzik U, Hupert-Kocurek K, Wojcieszyńska D (2015) Cometabolic degradation of naproxen by Planococcus sp. strain S5. Water Air Soil Pollut. https://doi.org/10.1007/s11270-015-2564-6
Fischer J, Kappelmeyer U, Kastner M et al (2010) The degradation of bisphenol A by the newly isolated bacterium Cupriavidus basilensis JF1 can be enhanced by biostimulation with phenol. Int Biodeterior Biodegradation 64:324–330. https://doi.org/10.1016/j.ibiod.2010.03.007
Fujiwara H, Soda S, Fujita M, Ike M (2016) Kinetics of bisphenol A degradation by Sphingomonas paucimobilis FJ-4. J Biosci Bioeng 122:341–344. https://doi.org/10.1016/j.jbiosc.2016.02.015
Furmanczyk EM, Kaminski MA, Lipinski L et al (2018) Pseudomonas laurylsulfatovorans sp. nov., sodium dodecyl sulfate degrading bacteria, isolated from the peaty soil of a wastewater treatment plant. Syst Appl Microbiol 41:348–354. https://doi.org/10.1016/j.syapm.2018.03.009
García-Espiñeira MC, Tejeda-Benítez LP, Olivero-Verbel J (2018) Toxic effects of bisphenol A, propyl paraben, and triclosan on caenorhabditis elegans. Int J Environ Res Public Health. https://doi.org/10.3390/ijerph15040684
Górny D, Guzik U, Hupert-Kocurek K, Wojcieszyńska D (2019) Naproxen ecotoxicity and biodegradation by Bacillus thuringiensis B1(2015b) strain. Ecotoxicol Environ Saf 167:505–512. https://doi.org/10.1016/j.ecoenv.2018.10.067
Grelska A, Noszczyńska M (2020) White rot fungi can be a promising tool for removal of bisphenol A, bisphenol S, and nonylphenol from wastewater. Environ Sci Pollut Res. https://doi.org/10.1007/s11356-020-10382-2
Heidari H, Sedighi M, Zamir SM, Shojaosadati SA (2017) Bisphenol A degradation by Ralstonia eutropha in the absence and presence of phenol. Int Biodeterior Biodegrad 119:37–42
Ike M, Chen MY, Jin CS, Fujita M (2002) Acute toxicity, mutagenicity, and estrogenicity of biodegradation products of bisphenol-A. Environ Toxicol 17:457–461. https://doi.org/10.1002/tox.10079
Juni E (2015) Bergey’s Manual of systematic of Archaea and Bacteria. Brisou and Prévot 1954, 727. https://doi.org/10.1002/9781118960608.gbm01203
Kamaraj M, Sivaraj R, Venckatesh R (2014) Biodegradation of Bisphenol A by the tolerant bacterial species isolated from coastal regions of Chennai, Tamil Nadu, India. Int Biodeterior Biodegradation 93:216–222. https://doi.org/10.1016/j.ibiod.2014.02.014
Kamaraj M, Rajeshwari S, Aravind J (2018) Isolation of Virgibacillus sp. strain KU4 from agricultural soil as a potential degrader of endocrine disruptor bisphenol-A. Int J Environ Sci Technol 15:2545–2550. https://doi.org/10.1007/s13762-017-1398-8
Kang JH, Kondo F (2002) Bisphenol A degradation by bacteria isolated from river water. Arch Environ Contam Toxicol 43:265–269. https://doi.org/10.1007/s00244-002-1209-0
Kolvenbach B, Schlaich N, Raoui Z, Prell J, Zühlke S, Schäffer A, Guengerich FP, Corvini PFX (2007) Degradation pathway of bisphenol A: Does ipso substitution apply to phenols containing a quaternary α-carbon structure in the para position? Appl Environ Microbiol 73 (15):4776–4784
Kolvenbach BA, Helbling DE, Kohler HPE, Corvini PFX (2014) Emerging chemicals and the evolution of biodegradation capacities and pathways in bacteria. Curr Opin Biotechnol 27:8–14. https://doi.org/10.1016/j.copbio.2013.08.017
Kozińska A, Paździor E, Pȩkala A, Niemczuk W (2014) Acinetobacter johnsonii and Acinetobacter lwoffii - The emerging fish pathogens. Bull Vet Inst Pulawy 58:193–199. https://doi.org/10.2478/bvip-2014-0029
Kumar S, Stecher G, Li M et al (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Larkin MA, Blackshields G, Brown NP et al (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Le TXH, Nguyen T, Van A, Yacouba Z et al (2017) Correlation between degradation pathway and toxicity of acetaminophen and its by-products by using the electro-Fenton process in aqueous media. Chemosphere 172:1–9. https://doi.org/10.1016/j.chemosphere.2016.12.060
Lei L, Aoyama I (2010) Effect-directed investigation and interactive effect of organic toxicants in landfill leachates combining Microtox test with RP-HPLC fractionation and GC/MS analysis. Ecotoxicology 19:1268–1276. https://doi.org/10.1007/s10646-010-0511-2
Li R, Liu Y, Chen G et al (2008) Physiological responses of the alga Cyclotella caspia to bisphenol A exposure. Bot Mar 51:360–369. https://doi.org/10.1515/BOT.2008.050
Li G, Zu L, Wong P et al (2012) Bioresource technology biodegradation and detoxification of bisphenol A with one newly-isolated strain Bacillus sp. GZB: kinetics, mechanism and estrogenic transition. Bioresour Technol 114:224–230. https://doi.org/10.1016/j.biortech.2012.03.067
Louati I, Dammak M, Nasri R et al (2019) Biodegradation and detoxification of bisphenol A by bacteria isolated from desert soils. 3 Biotech 9:1–11. https://doi.org/10.1007/s13205-019-1756-y
Marchlewicz A, Guzik U, Hupert-Kocurek K et al (2017) Toxicity and biodegradation of ibuprofen by Bacillus thuringiensis B1 (2015b). Environ Sci Pollut Res 24:7572–7584. https://doi.org/10.1007/s11356-017-8372-3
Mtibaà R, Olicón-Hernández DR, Pozo C et al (2018) Degradation of bisphenol A and acute toxicity reduction by different thermo-tolerant ascomycete strains isolated from arid soils. Ecotoxicol Environ Saf 156:87–96. https://doi.org/10.1016/j.ecoenv.2018.02.077
Murínová S, Dercová K (2014) Response mechanisms of bacterial degraders to environmental contaminants on the level of cell walls and cytoplasmic membrane. Int J Microbiol. https://doi.org/10.1155/2014/873081
Noszczyńska M, Piotrowska-Seget Z (2018) Bisphenols: application, occurrence, safety, and biodegradation mediated by bacterial communities in wastewater treatment plants and rivers. Chemosphere. https://doi.org/10.1016/j.chemosphere.2018.02.179
Okpokwasili GC, Nweke CO (2006) Microbial growth and substrate utilization kinetics. Afr J Biotechnol 5:305–317. https://doi.org/10.5897/AJB2006.000-5041
Pacwa-Płociniczak M, Płaza GA, Poliwoda A, Piotrowska-Seget Z (2014) Characterization of hydrocarbon-degrading and biosurfactant-producing Pseudomonas sp. P-1 strain as a potential tool for bioremediation of petroleum-contaminated soil. Environ Sci Pollut Res 21:9385–9395. https://doi.org/10.1007/s11356-014-2872-1
Pfeifer D, Chung YM, Hu MCT (2015) Effects of low-dose bisphenol A on DNA damage and proliferation of breast cells: the role of c-Myc. Environ Health Perspect 123:1271–1279. https://doi.org/10.1289/ehp.1409199
Plahuta M, Tišler T, Toman MJ, Pintar A (2014) Efficiency of advanced oxidation processes in lowering bisphenol A toxicity and oestrogenic activity in aqueous samples. Arh Hig Rada Toksikol 65:77–87. https://doi.org/10.2478/10004-1254-65-2014-2415
Płociniczak T, Chodór M, Pacwa-Płociniczak M, Piotrowska-Seget Z (2019) Metal-tolerant endophytic bacteria associated with Silene vulgaris support the Cd and Zn phytoextraction in non-host plants. Chemosphere 219:250–260. https://doi.org/10.1016/j.chemosphere.2018.12.018
Ramette A, Frapolli M, Saux MF, Le et al (2011) Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst Appl Microbiol 34:180–188. https://doi.org/10.1016/j.syapm.2010.10.005
Robinson JA, Tiedje JM (1983) Nonlinear estimation of monod growth kinetic parameters from a single substrate depletion curve. Appl Environ Microbiol 45:1453–1458. https://doi.org/10.1128/aem.45.5.1453-1458.1983
Roh H, Subramanya N, Zhao F, Yu C-P, Sandt J, Chu K-H (2009) Biodegradation potential of wastewater micropollutants by ammonia-oxidizing bacteria. Chemosphere 77 (8):1084–1089
Sarma H, Lee WY (2018) Bacteria enhanced lignocellulosic activated carbon for biofiltration of bisphenols in water. Environ Sci Pollut Res 25:17227–17239. https://doi.org/10.1007/s11356-018-2232-7
Sasaki M, Akahira A, Oshiman KI et al (2005a) Purification of cytochrome P450 and ferredoxin, involved in bisphenol A degradation, from Sphingomonas sp. strain AO1. Appl Environ Microbiol 71:8024–8030. https://doi.org/10.1128/AEM.71.12.8024-8030.2005
Sasaki M, Maki J, Oshiman K, Matsumura Y (2005b) Biodegradation of bisphenol A by cells and cell lysate from Sphingomonas sp. Biodegradation 449–459
Suyamud B, Inthorn D, Panyapinyopol B, Thiravetyan P (2018) Biodegradation of bisphenol A by a newly isolated Bacillus megaterium strain ISO-2 from a polycarbonate industrial wastewater. Water Air Soil Pollut. https://doi.org/10.1007/s11270-018-3983-y
Telke AA, Kalyani DC, Jadhav UU, Parshetti GK, Govindwar SP (2009) Purification and characterization of an extracellular laccase from a Pseudomonas sp. LBC1 and its application for the removal of bisphenol A. J Mol Catal B Enzym 61(3–4):252–260
Tišler T, Krel A, Gerželj U et al (2016) Hazard identification and risk characterization of bisphenols A, F and AF to aquatic organisms. Environ Pollut 212:472–479. https://doi.org/10.1016/j.envpol.2016.02.045
Toyama T, Sato Y, Inoue D et al (2009) Biodegradation of bisphenol A and bisphenol F in the rhizosphere sediment of Phragmites australis. J Biosci Bioeng 108:147–150. https://doi.org/10.1016/j.jbiosc.2009.03.011
Wolski EA, Murialdo SE, Gonzalez JF (2006) Effect of pH and inoculum size on pentachlorophenol degradation by Pseudomonas sp. Water SA 32:1–5. https://doi.org/10.4314/wsa.v32i1.5228
Xiong J, An T, Li G, Peng P (2017) Accelerated biodegradation of BPA in water-sediment microcosms with Bacillus sp. GZB and the associated bacterial community structure. Chemosphere 184:120–126
Zhao J, Chen X, Lin F et al (2014) Mechanism of toxicity formation and spatial distribution in activated sludge treating synthetic effluent containing bisphenol A (BPA). Chem Eng J 250:91–98. https://doi.org/10.1016/j.cej.2014.03.025
Zhou NA, Kjeldal H, Gough HL, Nielsen JL (2015) Identification of Putative Genes Involved in Bisphenol A Degradation Using Differential Protein Abundance Analysis of Sphingobium sp. BiD32. Environ Sci Technol 49:12231–12241. https://doi.org/10.1021/acs.est.5b02987
Zielinska M, Wojnowska-Baryla I, Cydzik-Kwiatkowska A (2019) Bisphenol A removal from water and wastewater. Bisphenol A removal from water and wastewater 1-115
Zühlke MK, Schlüter R, Henning AK et al (2016) A novel mechanism of conjugate formation of bisphenol A and its analogues by Bacillus amyloliquefaciens: detoxification and reduction of estrogenicity of bisphenols. Int Biodeterior Biodegrad 109:165–173. https://doi.org/10.1016/j.ibiod.2016.01.019
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Noszczyńska, M., Chodór, M., Jałowiecki, Ł. et al. A comprehensive study on bisphenol A degradation by newly isolated strains Acinetobacter sp. K1MN and Pseudomonas sp. BG12. Biodegradation 32, 1–15 (2021). https://doi.org/10.1007/s10532-020-09919-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-020-09919-6