Abstract
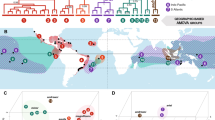
The largest anthropogenic extinction events during the Holocene occurred on Pacific islands, where thousands of bird populations were lost. Although ancient DNA approaches have become widely used to monitor the genetic variability of species through time, few studies have been conducted to identify the potential cryptic loss of genetic and species diversity within Pacific seabird species. Here we used heterochronous sampling of mitochondrial DNA (Cytochrome b) in the genus Pterodroma from Norfolk Island to quantify potential loss of genetic and species diversity. We particularly focused on the providence petrel P. solandri whose main breeding colony (~ 1,000,000 breeding pairs) became extirpated from Norfolk Island following European settlement circa 1800. We sampled subfossil bones consistent with Pterodroma spp. from Norfolk Island, and performed genetic comparisons with other populations of P. solandri and congeneric species. The majority of subfossil Norfolk Island individuals exhibited the most common mitochondrial haplotype from Lord Howe Island P. solandri, suggesting no appreciable loss of genetic variation as a consequence of the Norfolk Island extirpation. Our findings provide an example where a large seabird population was rapidly extirpated by humans without loss of species-level genetic diversity, probably as a consequence of high connectivity with other populations. However, past connectivity was insufficient to prevent the extirpation itself, which has conservation implications for predicting the resilience of threatened seabirds. In contrast, ancient DNA analyses of smaller Pterodroma bones from Norfolk Island indicate the loss of a second species, potentially P. pycrofti, P. brevipes or another closely related, possibly undescribed taxon, from the Tasman Sea.




Similar content being viewed by others
References
Anderson A, White JP (2001) The prehistoric archaeology of Norfolk Island, southwest Pacific. Records of the Australian Museum, supplement 27, Sydney
Bálint M, Domisch S, Engelhardt CHM et al (2011) Cryptic biodiversity loss linked to global climate change. Nat Clim Chang 1:313–318. https://doi.org/10.1038/nclimate1191
Beheregaray LB, Caccone A (2007) Cryptic biodiversity in a changing world. J Biol 6:1–5. https://doi.org/10.1186/jbiol60
Bester AJ (2003) The breeding, foraging ecology and conservation of the providence petrel, Pterodroma solandri, on Lord Howe Island, Australia. Unpublished Ph.D. Thesis, Charles Sturt University, Albury, NSW, Australia
Bester A, Klomp NI, Priddel D, Carlile N (2002) Chick-provisioning behaviour of the providence Petrel, Pterodroma solandri. Emu 102:297–304
Bickford D, Lohman DJ, Sodhi NS et al (2007) Cryptic species as a window on diversity and conservation. Trends Ecol Evol 22:148–155. https://doi.org/10.1016/j.tree.2006.11.004
Boessenkool S, Austin JJ, Worthy TH et al (2009) Relict or colonizer? Extinction and range expansion of penguins in southern New Zealand. Proc R Soc Lond B 276:815–821
Brodersen J, Seehausen O (2014) Why evolutionary biologists should get seriously involved in ecological monitoring and applied biodiversity assessment programs. Evol Appl 7:968–983. https://doi.org/10.1111/eva.12215
Brüniche-Olsen A, Jones ME, Burridge CP et al (2018) Ancient DNA tracks the mainland extinction and island survival of the Tasmanian devil. J Biogeogr 45:963–976. https://doi.org/10.1111/jbi.13214
Calvignac S, Hughes S, Tougard C et al (2008) Ancient DNA evidence for the loss of a highly divergent brown bear clade during historical times. Mol Ecol 17:1962–1970. https://doi.org/10.1111/j.1365-294X.2008.03631.x
Cascini M, Mitchell K, Cooper A, Phillips M (2018) Reconstructing the evolution of giant extinct kangaroos: comparing the utility of DNA, morphology, and total evidence. Syst Biol 68:520–537
Cooper A (2000) Ancient DNA: do it right or not at all. Science 289:1139–1139. https://doi.org/10.1126/science.289.5482.1139b
Cooper JH, Tennyson AJD (2008) Wrecks and residents: the fossil gadfly petrels (Pterodroma spp.) of the Chatham Islands. N Z Oryctos 7:227–248
Dabney J, Knapp M, Glocke I (2013) Complete mitochondrial genome sequence of a middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc Natl Acad Sci 110:15758–15763
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest2: more models, new heuristics and parallel computing. Nat Methods 9:772. https://doi.org/10.1038/nmeth.2109
Dellicour S, Mardulyn P (2014) SPADS 1.0: a toolbox to perform spatial analyses on DNA sequence data sets. Mol Ecol Resour 14:647–651
Eda M, Koike H, Kuro-o M et al (2011) Inferring the ancient population structure of the vulnerable albatross Phoebastria albatrus, combining ancient DNA, stable isotope, and morphometric analyses of archaeological samples. Conserv Genet 13:143–151. https://doi.org/10.1007/s10592-011-0270-5
Frankham R, Briscoe D, Ballou J (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Gamba C, Hanghøj K, Gaunitz C et al (2016) Comparing the performance of three ancient DNA extraction methods for high-throughput sequencing. Mol Ecol Resour 2:459–469. https://doi.org/10.1111/1755-0998.12470
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. https://doi.org/10.1080/10635150390235520
Heather B, Robertson H (1996) Field guide to the birds of New Zealand. Viking, Penguin Books Ltd, Auckland
Hermes N, Evans O, Evans B (1986) Norfolk Island birds: a review. Notornis 33:141–149
Hilton GM, Cuthbert RJ (2010) The catastrophic impact of invasive mammalian predators on birds of the UK Overseas Territories: a review and synthesis. Ibis 152:443–458
Hindwood K (1965) The “Sydney” bird paintings. Aust Zool 12:83–92
Hofreiter M, Serre D, Poinar HN et al (2001) Ancient DNA. Nat Rev Genet 2:353–359. https://doi.org/10.1038/35072071
Holdaway RN (1989) New Zealand’s pre-human avifauna and its vulnerability. N Z J Ecol 12:12–25
Holdaway RN, Anderson A (2001) Avifauna from the Emily Bay settlement site, Norfolk Island: a preliminary account. Rec Aust Museum 53:85–100. https://doi.org/10.3853/j.0812-7387.27.2001.1343
Holdaway RN, Worthy TH (1994) A new fossil species of shearwater puffinus from the late quaternary of the south island, New Zealand, and notes on the biogeography and evolution of the Puffinus gavia superspecies. Emu 94:201–215. https://doi.org/10.1071/MU9940201
Kirch P (1988) Polynesia’s mystery islands. Archaeology 41:26–31
Leigh JW, Bryant D (2015) POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116. https://doi.org/10.1111/2041-210X.12410
Leonard JA (2008) Ancient DNA applications for wildlife conservation. Mol Ecol 17:4186–4196
Lombal AJ, Wenner TJ, Carlile N et al (2017) Population genetic and behavioural variation of the two remaining colonies of providence petrel (Pterodroma solandri). Conserv Genet 18:117–129
Lowe WH, Allendorf FW (2010) What can genetics tell us about population connectivity? Mol Ecol 19:3038–3051. https://doi.org/10.1111/j.1365-294X.2010.04878.x
Medway DG (2002a) History and causes of the extirpation of the providence petrel (Pterodroma solandri) on Norfolk Island. Notornis 49:246–258
Medway DG (2002b) Why were providence petrels (Pterodroma solandri) nocturnal at Norfolk Island? Notornis 49:263–289
Meredith C (1985) The fossil fauna of Norfolk Island, South-west Pacific, and a review of the phylogeny of the genus Pterodroma. Unpublished PhD thesis, Monash University, Australia
Meyer M, Kircher M (2010) Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. https://doi.org/10.1101/pdb.prot5448
Milberg P, Tyrberg T (1993) Naïve birds and noble savages—a review of man-caused prehistoric extinctions of island birds. Ecography 16:229–250. https://doi.org/10.1111/j.1600-0587.1993.tb00213.x
Mitchell KJ, Wood JR, Scofield RP et al (2014) Ancient mitochondrial genome reveals unsuspected taxonomic affinity of the extinct Chatham duck (Pachyanas chathamica) and resolves divergence times for New Zealand and sub-Antarctic brown teals. Mol Phylogenet Evol 70:420–428. https://doi.org/10.1016/j.ympev.2013.08.017
Murphy SA, Joseph L, Burbidge AH, Austin J (2011) A cryptic and critically endangered species revealed by mitochondrial DNA analyses: the western ground parrot. Conserv Genet 12:595–600. https://doi.org/10.1007/s10592-010-0161-1
Niemiller ML, Graening GO, Fenolio DB et al (2013) Doomed before they are described? The need for conservation assessments of cryptic species complexes using an amblyopsid cavefish (Amblyopsidae: Typhlichthys) as a case study. Biodivers Conserv 22:1799–1820. https://doi.org/10.1007/s10531-013-0514-4
Orlando L, Cooper A (2014) Using ancient DNA to understand evolutionary and ecological processes. Annu Rev Ecol Evol Syst 45:573–598. https://doi.org/10.1146/annurev-ecolsys-120213-091712
Paxinos EE, James HF, Olson SL et al (2002) mtDNA from fossils reveals a radiation of Hawaiian geese recently derived from the Canada goose (Branta canadensis). Proc Natl Acad Sci 99:1399–1404. https://doi.org/10.1073/pnas.032166399
Pimm SL, Moulton MP, Justice LJ et al (1994) Bird extinctions in the central Pacific. Philos Trans R Soc Lond B 344:27–33. https://doi.org/10.1098/rstb.1994.0047
Ramakrishnan U, Hadly EA (2009) Using phylochronology to reveal cryptic population histories: review and synthesis of 29 ancient DNA studies. Mol Ecol 18:1310–1330. https://doi.org/10.1111/j.1365-294X.2009.04092.x
Ramírez O, Gómez-Díaz E, Olalde I et al (2013) Population connectivity buffers genetic diversity loss in a seabird. Front Zool 10:28
Rando JC (2002) New data of fossil birds from El Hierro (Canary Islands): probable causes of extinction and some biogeographical considerations. Ardeola 49:39–49
Rohland N, Harney E, Mallick S et al (2015) Partial uracil—DNA—glycosylase treatment for screening of ancient DNA. Philos Trans R Soc B 370:20130624. https://doi.org/10.1098/rstb.2013.0624
Saitoh T, Sugita N, Someya S et al (2015) DNA barcoding reveals 24 distinct lineages as cryptic bird species candidates in and around the Japanese Archipelago. Mol Ecol Resour 15:177–186. https://doi.org/10.1111/1755-0998.12282
Sala OE, Chapin FS, Armesto JJ et al (2000) Global biodiversity scenarios for the year 2100. Science 287:1770–1774. https://doi.org/10.1126/science.287.5459.1770
Schodde R, Fullagar P, Hermes N (1983) A review of Norfolk Island birds: past and present. Australian National Parks and Wildlife Service, Camberra
Schubert M, Ermini L, Sarkissian CD et al (2014) Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat Protoc 9:1056–1082. https://doi.org/10.1038/nprot.2014.063
Schwartz MK, Luikart G, Waples RS (2007) Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol 22:25–33
Smith AL, Friesen VL (2007) Differentiation of sympatric populations of the band-rumped storm-petrel in the Galápagos Islands: an examination of genetics, morphology, and vocalizations. Mol Ecol 16:1593–1603
Smith FDM, May RM, Pellew R et al (1993) Estimating extinction rates. Nature 364:494–496. https://doi.org/10.1038/364494b0
Steadman DW (1989) Extinction of birds in Eastern Polynesia: a review of the record, and comparisons with other Pacific Island groups. J Archaeol Sci 16:177–205. https://doi.org/10.1016/0305-4403(89)90065-4
Steadman DW (1995) Prehistoric extinctions of Pacific island birds: biodiversity meets zooarchaeology. Science 267:1123–1131. https://doi.org/10.1126/science.267.5201.1123
Steadman DW (2006) Extinction and biogeography of tropical Pacific birds. University of Chicago Press, USA
Steeves TE, Holdaway RN, Hale ML et al (2010) Merging ancient and modern DNA: extinct seabird taxon rediscovered in the North Tasman Sea. Biol Lett 6:94–97. https://doi.org/10.1098/rsbl.2009.0478
Tennyson AJD, Miskelly CM (2012) Observations of collared petrels (Pterodroma brevipes) on Vanua Lava, Vanuatu, and a review of the species’ breeding distribution. Notornis 59:39–48
Tennyson AJD, Cooper JH, Shepherd LD (2015) A new species of extinct Pterodroma petrel (Procellariiformes: Procellariidae) from the Chatham Islands, New Zealand. Bull Br Ornithol Club 135:267–276
Walker B (1996) Global change and terrestrial ecosystems. Cambridge University Press, UK
Warham J (1990) The Petrels: their ecology and breeding systems. A&C Black, UK
Welch AJ, Wiley AE, James HF et al (2012) Ancient DNA reveals genetic stability despite demographic decline: 3,000 years of population history in the endemic Hawaiian Petrel. Mol Biol Evol 29:3729–3740. https://doi.org/10.1093/molbev/mss185
Whitley G (1938) Naturalists of the first fleet. Aust Museum Mag 6:156–174
Wilmshurst JM, Moar NT, Wood JR et al (2014) Use of pollen and ancient DNA as conservation baselines for offshore islands in New Zealand. Conserv Biol 28:202–212. https://doi.org/10.1111/cobi.12150
Worthy TH, Holdaway RN (1993) Quaternary fossil faunas from caves in the Punakaiki area, west coast, South Island, New Zealand. J R Soc New Zeal 23:147–254. https://doi.org/10.1080/03036758.1993.10721222
Acknowledgements
We are grateful to David Binns, Norfolk Island Parks and Wildlife for providing logistics, transportation and accommodation during fieldwork on Norfolk Island. We thank the Museum of New Zealand Te Papa Tongarewa and the Australian National Wildlife Collections (ANWC) for sharing their collections of Pterodroma spp. from Norfolk Island and Lord Howe Island.
Funding
The SeaWorld Research and Rescue Foundation Inc (Grant SWR/4/2011) supported this work. Field collection of providence petrel blood samples was conducted with Animal Ethics permission from University of Tasmania Ethics Committee (Permit # A00011680).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Anurag chaurasia.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lombal, A.J., Salis, A.T., Mitchell, K.J. et al. Using ancient DNA to quantify losses of genetic and species diversity in seabirds: a case study of Pterodroma petrels from a Pacific island. Biodivers Conserv 29, 2361–2375 (2020). https://doi.org/10.1007/s10531-020-01978-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-020-01978-8