Abstract
We report the introduction of the central and eastern European helicid land snail Caucasotachea vindobonensis in North America. It was first recorded from Rensselaer County in the state of New York in 2015 by a community scientist. From 2016 to 2020, 14 additional occurrences in Rensselaer County, neighbouring Albany County and an imprecisely localized site in the Adirondack Mountains were recorded by community scientists. In 2020, the species was newly recorded at two sites in Schoharie County, NY, and at three sites in Québec, one of them approximately 700 km to the north of the initial record. Partial mitochondrial cox1 sequences from Rensselaer differ from an eastern Ukrainian haplotype only in a single substitution. Therefore, a Ukrainian origin for this introduction is likely, although not certain: the Rensselaer haplotype also differs in only two substitutions from a more widespread haplotype known from Ukraine, Hungary, Slovakia, Czechia, Serbia, and Bulgaria. An environmental niche model of the species based on occurrence data from central and eastern Europe indicated that a large region from the northern east coast to the midwestern United States is suitable for C. vindobonensis. The Canadian occurrences may indicate that the North American lineage is able to survive colder winters than predicted by the environmental niche model. Caucasotachea vindobonensis is not listed as a pest in Europe and it is unlikely to become an agricultural pest in North America as it prefers rotting plant material over living parts of plants, but its impact on native organisms can hardly be predicted.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Increasing international traffic and trade result in increasing numbers of human-mediated long-distance dispersals of animals and plants (Robinson 1999). The successful colonization of distant regions by invasive species adapted to anthropogenic habitats is further facilitated by the modification and homogenization of landscapes by humans. Such human-mediated dispersal is causing a breakdown of biogeographic barriers and thus climate is becoming more important for determining the composition of biotic communities, as has been shown especially for terrestrial snails (Capinha et al. 2015).
The land snail fauna of North America (north of Mexico) includes only about 1200 species (Nekola 2014), whereas 2480 species of land snails are known from Europe (exclusive of the Caucasus region; Neubert et al. 2019), although the area of North America is almost two times larger than that of Europe (19.8 million km2 versus 10.2 million km2, respectively). The colonization of North America by Europeans coupled with its comparatively depauperate native mollusc fauna may have favoured the introduction of a large number of European land snail species, especially those adapted to anthropogenic habitats. Gladstone et al. (2020) listed 69 introduced land snail species for which georeferenced occurrence data from the contiguous United States are available. The native ranges of 45 of these 69 introduced species are in Europe; eight additional species have a broad Mediterranean distribution that encompasses Western Europe and Northern Africa (Gladstone et al. 2020). In contrast, only 11 of the 2480 European land snail species are introduced (Neubert et al. 2019). The number of introduced land snail species in the contiguous United States increased approximately linearly from two around 1900 to the 69 currently recorded species (Gladstone et al. 2020). Recently, community science projects have also contributed to the documentation of newly introduced land snail species and their spread (Vendetti et al. 2018).
Here, we report another introduction to North America of a European land snail species, one first recorded and identified by community scientists. They found the central and eastern European helicid Caucasotachea vindobonensis (Pfeiffer, 1828) in the state of New York and in the Canadian province of Québec. We used mitochondrial DNA sequences to infer the probable region of origin of the introduction and we estimated the potential range of the species in North America using distribution modelling.
Materials and methods
Material, DNA extraction, PCR amplification and sequencing
After an initial identification of photographs of a snail from the state of New York as C. vindobonensis, several thousand observations on the community scientist platform iNaturalist (https://www.inaturalist.org/) from the northeastern United States and Ontario, Québec, and the Atlantic provinces of Canada were assessed.
For the molecular study, specimens of C. vindobonensis were collected in Rensselaer, at 100 New Broadway (42.6444°N 73.7419°W), in the state of New York and fixed in 95 % ethanol. Voucher specimens were deposited in the USDA APHIS National Mollusk Collection at the Academy of Natural Sciences in Philadelphia, USA (USDA 131,326, 5 specimens), and the Zoological Museum Hamburg, Germany (ZMH 140,788, 4 specimens).
Total genomic DNA was extracted from tissue samples following a slightly modified version of the protocol of Sokolov (2000) as detailed by Scheel and Hausdorf (2012). Parts of the mitochondrial cytochrome c oxidase subunit 1 gene (cox1) and the 16 S rDNA were amplified by polymerase chain reaction (PCR) using the primer pairs LCO1490 plus HCO2198 (Folmer et al. 1994) for cox1 and 16Scs1 plus 16Scs2 (Chiba 1999) for 16 S rDNA, respectively. Amplifications were performed in 25 µl volumes containing 2.5 µl 10× DreamTaq Green Buffer (Thermo Fisher Scientific, Waltham, MA, USA), 1 µl dNTP mix (5 mM each, biolabproducts, Bebensee, Germany), 1 µl of each primer (10 µM), 0.2 µl DreamTaq DNA polymerase (5 U/µl; Thermo Fisher Scientific), 0.5 µl template DNA and 19.8 µl ultrapure H2O. The reaction conditions were 95 °C for 2 min, 35 PCR cycles 95 °C for 30 s, 48 °C for 30 s, 72 °C for 1 min, followed by a final extension step at 72 °C for 7 min. Both strands of the amplified products were sequenced at Macrogen Europe (Amsterdam, The Netherlands) and assembled using ChromasPro 1.7.4 (Technelysium, South Brisbane, Australia).
Sequence alignment and phylogenetic analyses
We added the partial cox1 and 16 S rDNA sequences of the C. vindobonensis specimens from Rensselaer, New York (GenBank accession numbers MW292717, MW293719) to the alignment compiled by Korábek et al. (2020: Alignments S1). Only the 158 specimens from which cox1 as well as 16 S rDNA sequences were available were considered in the phylogenetic analysis.
The DNA sequences were initially divided into four partitions, the first, second, and third codon positions of cox1 and 16 S rDNA. An exhaustive search with PartitionFinder version 2.1.1 (Lanfear et al. 2017) was conducted to select an appropriate partitioning scheme and evolutionary models based on the corrected Akaike information criterion allowing for separate estimation of branch lengths for each partition. The models were limited to those available in MrBayes version 3.2.2 (Ronquist et al. 2012).
Bayesian inference of the phylogeny was performed using MrBayes version 3.2.2 (Ronquist et al. 2012). Metropolis coupled Markov chain Monte Carlo searches in MrBayes were run with four chains in two separate runs for 5,000,000 generations with default priors, trees sampled every 1,000 generations and separate estimation of parameters for individual partitions under default heating using best-fit models as suggested by PartitionFinder. Diagnostic tools provided in MrBayes were used to ensure that the runs had reached stationarity and convergence. The first 1,250,000 generations were discarded as burn-in.
We constructed a median-joining network (Bandelt et al. 1999) based on 292 partial cox1 sequences from the studies of Kajtoch et al. (2017) and Korábek et al. (2020) using the program PopART (Leigh and Bryant 2015) with ε = 0.
Environmental niche modelling
We compiled European records of C. vindobonensis from Kajtoch et al. (2017), Korábek et al. (2020), iNaturalist (https://www.inaturalist.org/observations?place_id=any&taxon_id=922180, accessed on 11.11.2020), and the material in the Zoological Museum Hamburg for environmental niche modelling (Electronic supplementary material 1). We used Maxent version 3.4.1 (Philips et al. 2020) for ecological niche modelling as implemented in the ‘kuenm’ workflow (Cobos et al. 2019). This workflow includes model calibration, model selection, and model evaluation of a pre-defined set of Maxent models. We assembled two data sets of 19 bioclimatic variables with a spatial resolution of 10 arc-minutes from the WorldClim 2.1 project (Fick and Hijmans 2017), one for Europe and another for North America, to which the best ecological niche model will be projected. Details of Environmental Niche Modelling approach are given in Electronic supplementary material 2.
Results
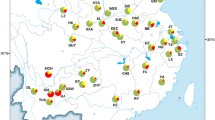
Caucasotachea vindobonensis in North America (Fig. 1a).
A photo of C. vindobonensis from a site next to the Hudson River in Troy, New York (Rensselaer County) was first uploaded in 2015 by a community scientist on the platform iNaturalist (https://www.inaturalist.org/observations/1537457). The photo shows several adult, bleached shells of C. vindobonensis indicating that a population was established already at that time. From 2016 to 2020, photographs of specimens from 14 additional localities in Troy and Rensselaer in Rensselaer County, in Cohoes and Albany in neighbouring Albany County, and a not precisely localized site in the Adirondack Mountains were uploaded, almost all of these before C. vindobonensis was identified in the region and recognized as a species of interest. With one observation each in the years 2015–2018, five in 2019, and 25 in 2020, observations of C. vindobonensis have been increasing at a rate that may simply reflect the rapidly increasing rate of community-science activity on the iNaturalist platform, but that may also indicate an increasing snail population (see https://www.inaturalist.org/observations/55864943). Following these observations in Rensselaer and Albany Counties, the species was recorded at two new sites near Howes Cave in neighbouring Schoharie County, and at three distant sites near Terrebonne and Dolbeau-Mistassi in Québec.
Origin of the American Caucasotachea vindobonensis
The mitochondrial cox1 and 16 S rDNA sequences of four specimens from Rensselaer in upstate New York proved to be identical. The alignment of concatenated cox1 (655 bp) and 16 S rDNA (823 bp) sequences of 158 C. vindobonensis has a total length of 1478 bp. PartitionFinder suggested using the following partitions and nucleotide substitution models: GTR + I for first codon positions of cox1, F81 for second codon positions of cox1, GTR + G for third codon positions of cox1, and GTR + I + G for 16 S rDNA. Bayesian inference analysis of the concatenated cox1 and 16 S rDNA sequences with these models showed that the haplotype from Rensselaer belongs to a clade of C. vindobonensis, which is widely distributed in Eastern Europe (Fig. 2).
The median-joining network based on 292 cox1 sequences indicated that the Rensselaer haplotype is most closely related to a haplotype from Krasnolimanskiy district in Ukraine, from which it differs only in a single substitution (Fig. 3). The American haplotype differs from a more widespread haplotype known from Ukraine and also from Hungary, Slovakia, Czechia, Serbia, and Bulgaria in two substitutions.
Potential distribution of Caucasotachea vindobonensis in North America
Details of the modelling results are given in Electronic supplementary material 2. The most important predictors for the occurrence of C. vindobonensis according to the jackknife test were Mean Temperature of Coldest Quarter (BIO11), Minimum Temperature of Coldest Month (BIO6), Mean Temperature of Wettest Quarter (BIO8), and Mean Temperature of Driest Quarter (BIO9). These variables are moderately correlated but they all point to minimum temperature as the most important limiting environmental parameter, and hence we interpret the boundary of the species to be driven by a lower temperature boundary.
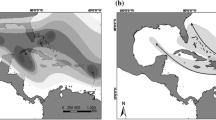
The environmental niche model for C. vindobonensis based on its distribution in Europe indicated that there are very suitable conditions for the species in a large region in the central and eastern United States (Fig. 1a). The species may also expand northwards along the Atlantic coast to Nova Scotia. More arid regions will probably be avoided by the species. The mobility-oriented parity analysis revealed that some parts of the projected areas with high probability should be interpreted with care such as the eastern areas of North and South Carolina as well as a predicted area near Roswell, New Mexico (Electronic supplementary material 3). The reported occurrences of C. vindobonensis in upstate New York are at the northern edge of the predicted suitable region, whereas the northernmost occurrences in Québec are outside of this predicted area.
Discussion
Caucasotachea vindobonensis is a central and eastern European species, which differs from the similar Cepaea species with which it is sometimes confused, by the finely ribbed, lighter-coloured shell with a light brownish lip (Fig. 3, insert). The species was actually included in Cepaea until recently, when molecular phylogenetics demonstrated that it is not related to western and central European Cepaea, but to Caucasian Caucasotachea (Korábek et al. 2015; Neiber and Hausdorf 2015; Neiber et al. 2016). Its natural range extends from the eastern rim of Germany, Poland, and an isolated occurrence in Latvia in the northwest, southwards through the Czech Republic, Austria, and northeastern Italy, across the Balkan Peninsula and southeastern Europe, with isolated occurrences in southern Russia eastwards to the Ural Mountains and from Crimea along the Caucasus to Dagestan (Electronic supplementary material 1; Neiber and Hausdorf 2015; Kajtoch et al. 2017; Korábek et al. 2020). Caucasotachea vindobonensis becomes adult within two years and copulates in spring and lays 29–67 eggs (mean of 49.3 ± 12.5) in their third year (Staikou 1998). Egg production decreases to 17.3 ± 3.8 eggs already in the fourth year. Caucasotachea vindobonensis may probably live up to 7 years (Staikou 1998). Staikou’s (1998) research at the southern margin of the species’ natural range in Greece suggested the capacity for population increase was low, due to a high mortality rate.
The occurrence of C. vindobonensis in North America, more precisely in the state of New York, was first documented in 2015. A population was already established at that time. Given the generation time of three years (Staikou 1998), the introduction to North America occurred at the beginning of the past decade at the latest. Caucasotachea vindobonensis has been recorded with increasing frequency from 14 sites in Rensselaer and Albany Counties. In 2020, the species was recorded for the first time from two sites in Schoharie County about 50 km west of the first occurrences. Finally, it was also recorded at three sites near Terrebonne and Dolbeau-Mistassi in Québec in Canada. These occurrences in Québec, north by around 300 and 700 km respectively of the New York populations, suggest either unexpectedly rapid human-mediated dispersal, establishment of C. vindobonensis in North America at a much earlier date than the present records show, or more than one introduction from Europe. The final scenario is one that has been proposed for Cepaea nemoralis in North America (Layton et al. 2018).
Molecular phylogenetic analysis of the available partial cox1 and 16 S rDNA sequences (Fig. 2) showed that the mitochondrial haplotype of C. vindobonensis from Rensselaer belongs to a widespread haplotype clade (Korábek et al. 2020: Fig. 2, yellow clade). A median-joining network based on 292 partial cox1 sequences indicated that the Rensselaer haplotype is most closely related to a haplotype from eastern Ukraine, from which it differs only in a single substitution (Fig. 3). Thus, a Ukrainian origin for the Rensselaer population is likely, although not certain: the Rensselaer haplotype also differs in only two substitutions from a more widespread haplotype known from a broader region including Ukraine, Hungary, Slovakia, Czechia, Serbia, and Bulgaria.
A projection of the environmental niche model based on occurrence data of C. vindobonensis from central and eastern Europe to North America (Fig. 1) indicated highly suitable climatic conditions for C. vindobonensis in a large area from the East coast to the Midwest of the United States. Thus, the species appears to have the potential to spread far west and southwards from its currently known range. The three Québec occurrences of the species reported here fall outside of the model’s predicted limits, a mismatch that will require further study. One possibility, considered unlikely, is that all three Québec individuals are of recent arrival and have yet to survive a winter or establish themselves. Another is that C. vindobonensis may be able to persist in generally colder regions where local microclimates are more favourable. Most of the North American populations observed are in close proximity to rivers or lakes, and to human settlements, both circumstances may provide microclimates that may mitigate cold weather extremes. A third possibility that cannot be excluded is that the North American lineage or lineages of C. vindobonensis are able to survive colder winters than predicted by the environmental niche model based on European occurrence data, for reasons still to be determined.
The risk that C. vindobonensis will become a significant agricultural pest in North America is low, as it prefers rotting plant material over living plant parts (Hatziioannou et al. 1994) and it is not considered a pest in Europe. However, it is possible that the species may become more widespread and abundant over time and given appropriate conditions. Given our current information it is hardly possible to predict the species’ potential negative impacts on native organisms and, if it becomes common enough, it may have a negative impact on agriculture and other land uses despite its preference for rotting plant material. Thus, its future spread and population development should continue to be monitored in North America.
The discovery of the introduction of C. vindobonensis in North America and the documentation of its spread demonstrates the increasing importance of community scientists in biodiversity monitoring efforts (Vendetti et al. 2018; Gladstone et al. 2020). The human resources gained with a community scientist platform such as iNaturalist can be essential to detect introduced species early, before they cause damage to the ecosystem, and to track their spatial distribution (Crall et al. 2010).
Availability of data and material
Voucher specimens were deposited in the USDA National Mollusk Collection at the Academy of Natural Sciences in Philadelphia, USA (USDA 131326), and the Zoological Museum Hamburg, Germany (ZMH 140788). DNA sequences were submitted to GenBank (accession numbers MW292717, MW293719).
Code availability
Not applicable.
References
Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Capinha C, Essl F, Seebens H, Moser D, Pereira HM (2015) The dispersal of alien species redefines biogeography in the Anthropocene. Science 348:1248–1251
Chiba S (1999) Accelerated evolution of land snails Mandarina in the oceanic Bonin Islands: evidence from mitochondrial DNA sequences. Evolution 53:460–471
Cobos ME, Peterson AT, Barve N, Osorio-Olvera L (2019) kuenm: An R package for detailed development of ecological niche models using Maxent. PeerJ 7:e6281
Crall AW, Newman GJ, Jarnevich CS, Stohlgren TJ, Waller DM, Graham J (2010) Improving and integrating data on invasive species collected by citizen scientists. Biol Invasions 12:3419–3428
Fick SE, Hijmans RJ (2017) WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int J Climatol 37:4302–4315
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Marine Biol Biotechnol 3:294–299
Gladstone NS, Bordeau TA, Leppanen C, McKinney ML (2020) Spatiotemporal patterns of non-native terrestrial gastropods in the contiguous United States. NeoBiota 57:133–152
Hatziioannou M, Eleutheriadis N, Lazaridou-Dimitriadou M (1994) Food preferences and dietary overlap by terrestrial snails in Logos area (Edessa, Macedonia, Northern Greece). J Molluscan Stud 60:331–341
Kajtoch Ł, Davison A, Grindon A, Deli T, Sramkó G, Gwardjan M, Kramarenko S, Mierzwa-Szymkowiak D, Ruta R, Ścibior R, Tóth JP, Wade C, Kolasa M, Egorov RV, Fehér Z (2017) Reconstructed historical distribution and phylogeography unravels non-steppic origin of Caucasotachea vindobonensis (Gastropoda: Helicidae). Org Divers Evol 17:679–692
Korábek O, Juřičková L, Petrusek A (2020) Inferring the sources of postglacial range expansion in two large European land snails. J Zool Syst Evol Res 58:944–956
Korábek O, Petrusek A, Neubert E, Juřičková L (2015) Molecular phylogeny of the genus Helix (Pulmonata: Helicidae). Zool Scripta 44:263–280
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2017) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol 34:772–773
Layton KKS, Warne CPK, Nicolai A, Ansart A, deWaard JR (2019) Molecular evidence for multiple introductions of the banded grove snail (Cepaea nemoralis) in North America. Canadian J Zool 97:392–398
Leigh JW, Bryant D (2015) popart: Full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116
Neiber MT, Hausdorf B (2015) Molecular phylogeny reveals the polyphyly of the snail genus Cepaea (Gastropoda: Helicidae). Mol Phylogen Evol 93:143–149
Neiber MT, Sagorny C, Hausdorf B (2016) Increasing the number of molecular markers resolves the phylogenetic relationship of ‘Cepaea’ vindobonensis (Pfeiffer, 1828) with the genus Caucasotachea Boettger, 1909 (Gastropoda: Pulmonata: Helicidae). J Zool Syst Evol Res 54:40–45
Nekola JC (2014) Overview of the North American terrestrial gastropod fauna. Amer Malac Bull 32:225–235
Neubert E, Seddon MB, Allen DJ, Arrébola J, Backeljau T, Balashov I, Bank R, Cameron R, de Frias Martins AM, De Mattia W, Dedov I, Duda M, Falkner G, Falkner M, Fehér Z, Gargominy O, Georgiev D, Giusti F, Gómez Moliner BJ, Groh K, Ibáñez M, Kappes H, Manganelli G, Martínez-Ortí A, Nardi G, Neiber MT, Páll-Gergely B, Parmakelis A, Prié V, Reischütz A, Reischütz PL, Rowson B, Rüetschi J, Slapnik R, Son M, Štamol V, Teixeira D, Triantis K, Vardinoyannis K, von Proschwitz T, Walther F (2019) European Red List of Terrestrial Molluscs. Snails, slugs, and semi-slugs. IUCN, Cambridge
Philips SJ, Dudík M, Schapire RE (2020) Maxent software for modeling species niches and distributions (version 3.4.1). http://biodiversityinformatics.amnh.org/open_source/maxent/
Robinson DG (1999) Alien invasions: the effects of the global economy on non-marine gastropod introductions into the United States. Malacologia 41:413–438
Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542
Scheel BM, Hausdorf B (2012) Survival and differentiation of subspecies of the land snail Charpentieria itala in mountain refuges in the Southern Alps. Mol Ecol 21:3794–3808
Sokolov EP (2000) An improved method for DNA isolation from mucopolysaccharide-rich molluscan tissues. J Molluscan Stud 66:573–575
Staikou AE (1998) Aspects of life cycle, population dynamics, growth and secondary production of the pulmonate snail Cepaea vindobonensis (Férussac, 1821) in northern Greece. J Molluscan Stud 64:297–308
Vendetti JE, Lee C, LaFollette P (2018) Five new records of introduced terrestrial gastropods in southern California discovered by citizen science. Am Malacol Bull 36:232–247
Acknowledgements
We are grateful to all of the iNaturalist community scientists who have documented the introduction and establishment of C. vindobonensis in North America, namely S. Cox, B. Dagley, S. Hankamp, S. Hewitt, K. Huang, E. Irons, R. Kelchlin, O. Kellhammer, J. Kent, B. Renaud, Z. Schwartz-Weinstein, G. Strano, J. Vanderwall, E. Wilmot, and other iNaturalists. We especially thank E. Caruso for collecting specimens for molecular analyses and J. Lauschke for performing DNA extraction and PCR and making the photo of the shell.
Funding
Open Access funding enabled and organized by Projekt DEAL. We received no funding for this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
All authors agreed to publish the manuscript.
Consent to participate
All authors agreed to participate in the study and its publication.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hausdorf, B., Parr, M., Shappell, L.J. et al. The introduction of the European Caucasotachea vindobonensis (Gastropoda: Helicidae) in North America, its origin and its potential range. Biol Invasions 23, 3281–3289 (2021). https://doi.org/10.1007/s10530-021-02579-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-021-02579-4