Abstract
Objectives
To investigate the effect of endogenous Cas9 on genome editing efficiency in transgenic zebrafish.
Results
Here we have constructed a transgenic zebrafish strain that can be screened by pigment deficiency. Compared with the traditional CRISPR injection method, the transgenic zebrafish can improve the efficiency of genome editing significantly. At the same time, we first observed that the phenotype of vertebral malformation in early embryonic development of zebrafish after ZFERV knockout.
Conclusions
The transgenic zebrafish with expressed Cas9, is more efficient in genome editing. And the results of ZFERV knockout indicated that ERV may affect the vertebral development by Notch1/Delta D signal pathway.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Zebrafish are a classical animal model for developmental biology and biomedical studies; several important questions in vertebrate development have been addressed using this model. Particularly, large-scale screening in the 1990s for zebrafish mutants generated via random mutagenesis resulted in zebrafish becoming one of the most frequently used animal models.
In recent years, newly developed genome-editing tools, including ZFNs, TALENs, and CRISPR/Cas9, provide a feasible approach for the generation of zebrafish with precise genomic modifications. These zebrafish have significantly promoted the understanding of genes with unknown functions, especially those involved in embryonic development (Jao et al. 2013). Specifically, CRISPR/Cas9 has become a popular method for site-specific genome editing due to its convenience and efficiency. However, as fertilized zebrafish eggs develop quickly, direct delivery of large-sized Cas9 mRNA or protein into eggs frequently poses the problem of chimerism. Generally, establishing a zebrafish homozygous line with a genomic modification from the mosaic F0 generation of zebrafish is time consuming, requiring breeding and genotyping of the F1 and F2 generations (Varshney et al. 2015). Thus, generating homozygous zebrafish with genomic modifications at the F0 generation would be very efficient and useful. Transgenic Cas9 zebrafish may provide an opportunity to meet this requirement.
In addition, transgenic Cas9 zebrafish may facilitate the generation of zebrafish with the deletion of a large genomic fragment. The CRISPR/Cas9 system has been used to knock out long non-coding RNAs that are usually large in size (Wen et al. 2016), as well as other large genomic DNA sequences (Kraft et al. 2015). Endogenous retroviruses (ERVs), which might be remnants of ancient retroviral infections, are large genomic elements. Zebrafish possess an 11349-bp ERV on chromosome 19 that contains the Gag, Pol, and Env genes, as well as long terminal repeat (LTR) sequences (Shi et al. 2015). Previous studies have shown that ERVs play an important role in the activation of innate immunity, embryonic development and the occurrence of diseases and tumors (Chuong et al. 2016). Given that zebrafish embryos can develop in vitro and because the effects of ERV deletion can be observed during embryonic development, ERV-deficient zebrafish may serve as a good model for ERV studies.
In this study, a Cas9 transgenic zebrafish line with a selective marker for homozygosity was successfully established, solving the problem of chimerism that occurs following the injection of large-sized Cas9 mRNA or protein into fertilized zebrafish eggs. Moreover, the generation of ERV-deficient zebrafish using the Cas9 transgenic zebrafish line indicates that the transgenic Cas9 gene is active and can be used to modify any gene of interest in zebrafish. Finally, ERV-deficient zebrafish may serve as animal models for ERV studies, and the spinal abnormality observed in ERV-deficient fish was associated with the downregulation of the delta D and Notch 1 genes.
Materials and methods
Maintenance and breeding of zebrafish
TU wild-type zebrafish (Danio rerio) were obtained from China Zebrafish Resource Center or CZRC (Wuhan, China). The fish were maintained at 28.5 °C under a 14 h light: 10 h dark–light regime (i.e., 14hL: 10hD) in the Zebrafish Research Facility, Yangzhou University (Yangzhou, China). To generate offspring, mating was carried out at a ratio of 1 female to 2 males, followed by natural spawning. Embryos were raised in E3 medium (5 mmol/L NaCl, 0.17 mmol/L KCl, 0.33 mmol/L CaCl2, and 0.33 mmol/L MgSO4, pH 7.2) at 28.5 °C on a 14 h light:10 h dark cycle. All animal experiments were performed in accordance with the guidelines for animal welfare in China, and the animal protocol was approved by the animal welfare committee of Yangzhou University.
Acquisition of zebrafish endogenous promoters and construction of eGFP expression plasmids
The zebrafish Eef1g (eukaryotic translation elongation factor 1 gamma) (Burket et al. 2008) and Ef1α (elongation factor 1) (Moon et al. 2013) promoter sequences were PCR-amplified using the primers specific to the promoters (Supplemental Table 1) and genomic DNA samples isolated from zebrafish. To link the promoters to the eGFP expression plasmid, the promoter sequences were further PCR-amplified using primers containing MluI (New England Biolabs (NEB), USA, Catalog no. R3198S) and NheI (NEB, USA, Catalog no. R3131S) restriction sites. The eGFP fragments were obtained by amplifying the pIRES2-eGFP plasmid using the primers eGFP-BamHI-F and eGFP-XhoI-R (Supplemental Table 1). The fragment was inserted into the pcDNA3.1 plasmid by digesting the plasmids with BamHI (NEB, USA, Catalog no. R3136S) and XhoI (NEB, USA, Catalog no. R0146S), followed by ligation with T4 ligase according to the manufacturers’ instructions. The map of the eGFP expression vector is shown in Supplemental Fig. 1. To generate eGFP constructs driven by the zebrafish Eef1g or Ef1α promoter, the CMV promoter was removed from the pcDNA3.1-eGFP plasmid by digestion with MluI and NheI (Supplemental Fig. 2), followed by ligation with the zebrafish Eef1g or Ef1a promoter using T4 ligase according to the manufacturers’ instructions. The resulting pcDNA3.1 plasmids containing Ef1α-eGFP or Eef1g-eGFP fragments are shown in Supplemental Figs. 3 and 4.
Microinjection of eGFP expression cassettes driven by zebrafish promoters
To generate zebrafish with Eef1g-eGFP or Ef1α-eGFP expression cassettes, microinjection solutions were prepared containing 100 ng Eef1g-eGFP (i.e., pcDNA3.1 [Eef1g-eGFP]) or Ef1α-eGFP (i.e., pcDNA3.1 [Ef1α-eGFP]) plasmid, 1% phenol red indicator, and water up to 5 µL (Supplemental Fig. 1). pcDNA3.1 [eGFP] plasmid containing eGFP and no promoter was used as a control (Supplemental Fig. 1). Microinjection was performed using a microinjector (TRITECH™ RESEARCH microINJECTOR system, Veteran Avenue, Los Angeles, USA, Catalog no. MINJ-1) under an inverted microscope (Zebrasc, Beijing, China) as described previously (Rosen et al. 2009). Each fertilized zebrafish egg was injected with 1 nL of each solution at the one or two-cell stage, and the injected embryos were subsequently raised at 28 °C in E3 medium. The fluorescent signal from the injected embryos was examined on a fluorescence microscope (Olympus FluoView IX73, Japan, Catalog No. IX7301F). The promoter activity was evaluated based on fluorescence emission.
Construction of Cas9 expression cassettes
The DNA sequence of the Cas9 gene was synthesized from the MLM3613 plasmid (Addgene, Catalog no. 42251). For the construction of the Cas9 expression cassette, the Ef1α promoter was inserted into MLM3613 plasmid via Sep I (NEB, Catalog No. R3133S) and NheI digestion, followed by ligation with T4 ligase according to the manufacturers’ instructions. The expression cassette consists of the following elements: Ef1α-Cas9-NLS-bGH poly (A). For locus-specific integration of the Cas9 expression cassette at the Mitfa locus in the zebrafish genome, Genewiz, Inc. (Suzhou, China), synthesized the 40-bp-long homologous recombination arms (HRL and HRR) around the Mitfα sgRNA identification site, together with the linker primers Cas9 cassette-F and Cas9 cassette-R (Supplemental Table 1). The purified product was used in PCR with the aforementioned Cas9 expression cassette to produce the Cas9 transgenic cassette: HRL-Ef1α-Cas9-NLS-bGH poly (A)-HRR. The PCR procedure included initial denaturation at 92 °C for 2 min, followed by 30 cycles of 92 °C for 30 s, 62 °C for 2 min 30 s, and 72 °C for 10 min, which was based on Vazyme’s long chain PCR settings (Vazyme, China, Catalog no. P501-d1). The final PCR product was isolated and purified using the Gel Recovery Kit (OMEGA, USA, Catalog no. D2500-02) according to the manufacturer’s instructions.
In vitro sgRNA and Cas9 mRNA synthesis
sgRNAs for Mitfα, Tyr and zebrafish ERV (ZFERV) were designed by Chop–Chop (Labun et al. 2016). The DNA templates for in vitro sgRNA synthesis, which contained the T7 promoter, a 20-nucleotide (nt) long guide sequence, and an sgRNA scaffold (Chen et al. 2013), were generated by overlapping PCR (Lin et al. 2014). PCR was performed using Phanta™® Super-Fidelity DNA polymerase (Vazyme, Catalog no. P501-d1) and the annealed product of two oligos (Supplemental Table 1) (Varshney et al. 2015). The annealing and PCR reactions contained 5 µL of 5× Super-Fidelity (SF) Buffer, 1 µL of dNTP Mix, 1 µL each of 10 µM oligos, 1 µL of Phanta® Super-Fidelity DNA Polymerase and 16 µL of water. The PCR settings were the same as those described previously (Chen et al. 2016). The sgRNA sequences are shown in Table 2. In vitro sgRNA transcription was carried out in reactions containing 1 µg of DNA template, 2 µL of 10× reaction buffer, 2 µL of ATP (100 mM), 2 µL of GTP (100 mM), 2 µL of UTP (100 mM), 2 µL of CTP (100 mM), 2 µL of T7 RNA Polymerase Mix (HiScribe™ T7 High Yield RNA Synthesis Kit, NEB, Catalog no. E2040S), and water up to 20 µL. In vitro transcription was performed by incubation at 37 °C for more than 16 h, followed by treatment with RNase-free DNase I (NEB, Catalog no. M0303S) at room temperature for 30 min. The synthesized sgRNAs were purified by the phenol–chloroform method and dissolved in pure water. To acquire Cas9 mRNA, Cas9 DNA templates from the MLM3613 plasmids were used for in vitro transcription. The templates were acquired by digesting the MLM3613 plasmid with PmeI (NEB, Catalog no. R0560S) and were purified using the Cycle Pure Kit (OMEGA, Catalog no. D6492-02) according to the manufacturers’ instructions. In vitro transcription reactions contained 1 µg of DNA template, 10 µL of 2× NTP/CAP, 2 µL of 10× reaction buffer, 2 µL of T7 RNA polymerase (mMESSAGE mMACHINE™ T7 Transcription Kit, Thermo Fisher, Catalog no. AM1344), and water up to 20 µL. The settings for in vitro transcription included incubation at 37 °C overnight, followed by treatment with RNase-free DNase I at room temperature for 30 min. The synthesized Cas9 mRNAs were purified by the phenol–chloroform method and dissolved in pure water. All synthesized RNA products were stored at -80 °C before microinjection.
Generation of Cas9 transgenic zebrafish
To generate zebrafish containing the Cas9 expression cassette, microinjection solutions were prepared containing 1000 ng of Cas9 mRNA, 200 ng of Mitfα sgRNA, 500 ng of Cas9 donor fragment, 1% phenol red, and water up to 5 µL. Fertilized zebrafish eggs were injected with 1 nL of solution, and embryos were raised at 28 °C in E3 medium.
Genotyping and sequencing analysis of transgenic zebrafish
For genotyping, genomic DNA was extracted from the caudal fins of 1-month-old fries, as described previously (Liu and Zhang 2011). Genotyping was performed by PCR using the gene-specific primers Cas9-test-F and Cas9-test-R (Supplemental Table 1), followed by sequencing analysis. To validate the integration of the Cas9 gene at the Mitfα locus, PCR was performed with a pair of primers that recognize Cas9 or Mitfα gene sequences. The 50-µL PCR reaction contained 10 µL of 5× SF Buffer, 1 µL of dNTP Mix, 2 µL of Cas9 insertion-test-F, 2 µL of Cas9 insertion-test-R, 1 µL of Phanta® Super-Fidelity DNA Polymerase, and 34 µL of water. The PCR settings included denaturation at 94 °C for 5 min, followed by 30 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min, and 72 °C for 10 min for final extension. The purified PCR product was subjected to sequencing analysis by Genewiz, Inc.
RT-PCR-based evaluation of transgenic Cas9 gene expression in the F2 generation
To determine the expression of the transgenic Cas9 gene, the total RNA was isolated from F2 zebrafish embryos with the Nacre phenotype using TRIzol (Ambion, Catalog no.15596018) according to the manufacturer’s instructions. Complementary DNA was synthesized using the total RNA samples and the PrimeScript™ 1st Strand cDNA Synthesis Kit (TAKARA, Catalog no.6110B) according to the manufacturer’s instructions. PCR was subsequently conducted with the cDNA samples and the primers Cas9-test-F and Cas9-test-R (Supplemental Table 1). Additionally, mock reverse transcription with the total RNA was performed as a negative control (no reverse transcriptase added), while PCR using the genomic DNA isolated from the F2 zebrafish was used as a positive control. All PCR products were separated via 1% agarose gel electrophoresis.
Generation of zebrafish with a Tyr gene mutation using Cas9 mRNA or transgenic Cas9
To generate Tyr-mutant zebrafish, two solutions for microinjection were prepared. The first solution, injected into Cas9 transgenic fish, contained 200 ng of Tyr sgRNA, 1% phenol red, and water up to 5 µL. The second solution, injected into wide-type TU fish, contained 1000 ng of Cas9 mRNA, 200 ng of Tyr sgRNA, 1% phenol red, and water up to 5 µL. Embryos were injected with 1 nL of solution. All embryos were maintained at 28 °C for at least one day before viability evaluation.
T7E1 mutagenesis assay of the Tyr gene
The T7E1 assay was performed as described previously (Kim et al. 2009). Genomic DNA was isolated from control TU zebrafish, those developed from Cas9 transgenic zebrafish fertilized eggs injected with Tyr sgRNA, and those developed from TU zebrafish fertilized eggs injected with Cas9 mRNA and Tyr sgRNA. A general PCR was performed with the genomic DNA samples and the primers Tyr-test-F and Tyr-test-R (Supplemental Table 1). After gel purification, 200 ng of purified DNA was reannealed in a thermal cycler under the following conditions: 95 °C for 2 min, 95 °C to 85 °C at a rate of 2 °C/s and 85 °C to 25 °C at a rate of 0.1 °C/s. Next, 16 µL of the reannealed product was incubated in a reaction solution containing 0.2 µL of T7 endonuclease I (NEB, USA, Catalog no. M0302S), 2 µL of NEB buffer 2, and 1.8 µL of nuclease-free water at 37 °C for 40 min. The digestion product was separated via 1.2% agarose gel electrophoresis.
Generation of ZFERV-deficient zebrafish
To generate ZFERV-deficient zebrafish and to compare the efficiency of transgenic Cas9 and Cas9 mRNA for genome editing, two microinjection solutions were prepared. One, injected into Cas9 transgenic fish, contained 200 ng of LTR sgRNA, 1% phenol red, and water up to 5 µL. The second, injected into TU fish, contained 1000 ng of Cas9 mRNA, 200 ng of LTR sgRNA, 1% phenol red, and water up to 5 µL. A total of 1 nL of each solution was used for microinjection. All embryos were raised at 28 °C in the E3 medium.
Detection of ZFERV deficiency
To confirm the ZFERV deletion, genomic DNA was extracted from zebrafish embryos after 24 h of development, from embryos with spinal deformity after 48 h of development, and from embryos with spinal deformity after 120 h of development. PCR was performed with the genomic DNA samples and each of the following primers pairs: for detection of the ZFERV 5′-LTR sequence, 5′LTR-test-F and 5′LTR-test-R (Supplemental Table 1); for detection of the ZFERV full-length sequence, 5′LTR-test-F and 3′LTR-test-R (Supplemental Table 1). The PCR solution contained 2 µL of genomic DNA, 2 µL of primers (1 µL each), 1 µL of Phanta® Super-Fidelity DNA Polymerase, 4 µL of 5× SF Buffer, 1 µL of dNTPs, and 10 µL of water. The PCR settings included denaturation at 94 °C for 10 min, followed by 30 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s, and 72 °C for 5 min for final extension. The PCR product was separated via 1% agarose gel electrophoresis and subjected to sequencing analysis by Genewiz, Inc.
qPCR analysis with the ZFERV-deficient zebrafishes
To determine Env, Delta D, Notch 1 and Her1 gene expression, total RNA was isolated using TRIzol according to the manufacturer’s instruction. Quantitative PCR was performed on a CFX Connect™ Real-time System (Bio-Rad, Serial no. 788BR04566). The reaction solutions contained 10 µL of 2× SYBR®Premix ExTaq (Clontech, Japan, Catalog no. 639676), 0.8 µL of Forward Primer (10 µm), 0.8 µL of Reverse Primer (10 µm), 2 µL of cDNA, and water up to 20 µL. The qPCR settings included denaturation at 95 °C for 3 min, followed by 40 cycles of 95 °C for 10 s and 58 °C for 30 s. The primers for the four genes and internal control genes Gapdh and Ef1α are listed in Supplemental Table 1. Relative gene expression levels were calculated with the 2−∆∆CT method, as indicated in the manufacturer guidelines (Bio-Rad).
Statistical analysis
One-way analysis of variance (ANOVA) was performed to determine the statistical significance of the difference in the expression of a given gene among different treatments. The level of statistical significance was set at p value < 0.05. All data are presented as the mean ± SE.
Results and discussion
Selection of the promoter to drive expression of the transgene in zebrafish
To efficiently and constantly drive the expression of exogenous Cas9 gene in zebrafish, two candidate endogenous zebrafish promoters, Eef1g (eukaryotic translation elongation factor 1 gamma) and Ef1α (elongation factor 1), were used in the transgenic study. Both promoters are known to drive genes expressed during early zebrafish development. Using zebrafish genomic DNA as a PCR template, Ef1α (942 bp) and Eef1g promoter sequences (2531 bp) were successfully amplified (Fig. 1a) and confirmed by sequence analysis of the PCR products. The promoter sequences were separately cloned into expression vectors to drive the expression of a green fluorescent protein reporter gene (Fig. 1b). In total, 482 of 642 zebrafish (76%) in the Ef1α group were fluorescent, while 400 of 540 zebrafish (74%) in the Eef1g group were fluorescent, suggesting that the two promoters were able to drive reporter gene expression with similarly efficiency. The Ef1α promoter is much shorter than the Eef1g promoter and may also possess better integration efficiency; we thus chose the Ef1α promoter as the promoter for the Cas9 gene to construct the transgenic donor fragment.
Identification of Cas9 transgenic zebrafish
Using genomic DNA isolated from the fins of F0 zebrafish developed from injected eggs, Cas9 was PCR-amplified using specific primers, indicating that the Cas9 gene was successfully integrated into the zebrafish genome via the CRISPR-Cas9 system (119/403, approximately 30% of F0 zebrafish). Amplicons with the expected size of 453 bp were sequenced and confirmed (Fig. 2b). To confirm if the integration of the Cas9 gene was correctly targeted to the Mitfα locus, a pair of primers (Cas9 insertion-test-F and Cas9 insertion-test-R) was designed to be specific to both the Cas9 gene and the flanking Mitfα sequence. A PCR amplicon with of the expected size (1383 bp) was obtained (Fig. 2c), and subsequent sequencing analysis confirmed that the Cas9 gene was specifically inserted into the Mitfα locus. To evaluate the activity of the inserted Cas9 gene, zebrafish that developed from the injected eggs were used to generate the next generation of zebrafish with biallelic disruption of the Mitfα gene. As only fish with biallelic Mitfα disruption would exhibit a distinct phenotype from those with monoallelic or no Mitfα disruption in terms of body pigmentation (Fig. 2d), offspring with biallelic-mutation Mitfα were easily selected by appearance (260 out of 1130 or 23%). Transgenic Cas9 expression gene was detected in these offspring (Fig. 2e), suggesting that Cas9 was constitutively expressed in the transgenic zebrafish line (Fig. 3a). Using this line, sgRNA targeted to Tyr was injected to generate Tyr-mutant zebrafishes. T7E1 restriction enzyme digestion showed that Tyr was successfully mutated. The results also indicated that the editing efficiency of transgenic Cas9 was better than that of exogenous Cas9 mRNA (Fig. 3b), and the phenotypic change caused by Tyr deletion is shown in Fig. 3c. These experiments confirmed that the transgenic Cas9 functioned efficiently.
Generation of Cas9 transgenic zebrafish. a Schematic illustration of Mitfα locus-specific integration of the Ef1α-Cas9 expression cassette based on homologous recombination using Cas9 mRNA and sgRNA targeted to the zebrafish Mitfα gene. The indicated primers were used to confirm integration. HR denotes homologous recombination arm, and NLS denotes nuclear localization signal. b Confirmation of Cas9 gene insertion into the zebrafish genome, indicated by amplification of Cas9 by PCR with Cas9-specific primers. The expected size of the Cas9 amplicons is 453 bp. TU denotes genomic DNA isolated from wild-type zebrafish, while Cas9 denotes genomic DNA isolated from Cas9 transgenic zebrafish. M denotes the DNA ladder. c Confirmation of Mitfα-locus-specific integration of the Cas9 gene into the zebrafish genome, indicated by amplification of Cas9 by PCR with both Mitfα- and Cas9-specific primers. The expected size of the Cas9 amplicons is 1383 bp. TU denotes genomic DNA isolated from wild-type zebrafish, while Cas9 denotes genomic DNA isolated from Cas9 transgenic zebrafish. M denotes the DNA ladder. d Selection of zebrafish with biallelic transgenic Cas9 gene based on the nacre phenotype (pigment-deficient mutant), including genotyping and breeding of F0 zebrafish and selection for nacre phenotype-carrying zebrafish in F1. Based on phenotype, 23% (260/1130) of F1 zebrafish carry the nacre phenotype. The red box denotes the transgenic Cas9 gene. e Comparison of phenotypes of TU wild-type zebrafish with those of Mitfα-locus-specific transgenic Cas9 zebrafish at different developmental stages. Hpf and mpf indicate hours post-fertilization and months post-fertilization, respectively
Validation of Cas9 activity in F2 transgenic zebrafish. a Transgenic Cas9 expression was detected by reverse transcription (RT)-PCR in F2 transgenic zebrafish. Lane G: genomic DNA from F2 transgenic zebrafish as the positive control. Lane RT-: RT with no reverse transcriptase as the negative control. Lane RT + : cDNA synthesized by RT with total RNA isolated from F2 transgenic zebrafish. Lane M: DNA ladder. The expected size of the amplicon is 453 bp. b Validation of transgenic Cas9 activity by Tyr mutation assay and comparison of the editing efficiency of exogenous Cas9 mRNA and transgenic Cas9. The top panel shows the targeting site of the Tyr sgRNA in the Tyr gene, as well as the binding sites of the PCR primers used to detect the Tyr mutation in F2 zebrafish. The bottom panel shows the detection of the Tyr mutation via PCR and T7E1 digestion in fertilized eggs from TU wild-type zebrafish, which were injected with Tyr sgRNA plus Cas9 mRNA (a) vs. fertilized eggs from Cas9 transgenic zebrafish, which were injected with Tyr sgRNA (b), and TU wildtype zebrafish as control (c). The Tyr mutation was indicated by two fragments (approximately 550 bp and 500 bp, as indicated). c Different phenotypic changes in pigmentation fading in Tyr knockout zebrafish embryos produced via different gene editing methods. The representative images show a embryos injected with Cas9 mRNA and sgRNA, b Cas9 transgenic embryos injected with sgRNA and c wild-type embryos
ERV deletion causes spinal abnormality in zebrafish
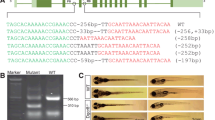
Recent studies indicate that ERVs play a role in humans and other animals. As an animal model of ERV knockout is urgently needed, the transgenic Cas9 zebrafish line was used to generate an ERV-knockout model. An sgRNA targeting the long terminal repeat (LTR) of zebrafish ERV (ZFERV, 11349 bp in length) was designed (Fig. 4a). After injection, embryos were collected at different stages, and genomic DNA was then extracted for PCR. The primers 5′LTR-test-F, 5′LTR-test-R were used to amplify the 5′LTR sequence around the sgRNA-targeted site. As expected, no amplicons were observed in the ERV-knockout fish due to the removal of the partial 5′LTR sequence that contains the binding site for the reverse primer (Supplemental Fig. 5), whereas a 284-bp PCR product was detected in wild-type fish (Fig. 4b). Consistently, using the primers 5′LTR-test-F, 3′LTR-test-R, which recognize the 5′LTR and 3′LTR sequences, respectively, a 609-bp amplicon was obtained in the ERV-knockout fish, while the expected 11182-bp fragment from wild-type fish was too large to amplify (Fig. 4c). These results suggested that ERV was successfully knocked out with the Cas9 transgenic zebrafish. Sequencing analysis further confirmed that ERV was removed from the zebrafish. Surprisingly, most of the embryos (2271 out of 2640, or 86%) showed spinal abnormalities 48 h after injection (Fig. 4d). Zebrafish embryos with abnormal spines had difficulty swimming.
Generation of ERV-deficient zebrafish. a Schematic overview of ZFERV gene knockout. 5′LTR-test-F and 5′LTR-test-R primers were used to PCR-amplify the 5′LTR sequence, while 5′LTR-test-F and 3′LTR-test-R primers were used to PCR-amplify the sequence between the 5′ and 3′LTR. b Gel images of PCR amplicons synthesized using GAPDH primers and the 5′LTR-test-F and 5′LTR-test-R primers for the ZFERV 5‘LTR used in the KO test. The primers 5′LTR-test-F and 5′LTR-test-R were used to amplify the ZFERV 5′LTR sequence. The expected sizes of the amplicons are indicated. Lane a: collected fertilized zebrafish eggs injected with LTR sgRNAs; Lane b and Lane c: zebrafish embryos with spinal deformity at 48 h and 120 h post-fertilization; Lane d: TU wild-type zebrafish embryos; M denotes the DNA ladder. c Detection of ZFERV gene deletion, which is indicated by the gel image of PCR amplicons synthesized from zebrafish genomic DNA using the primers 5′LTR-test-F and 3′LTR-test-R. The primers 5′LTR-test-F and 3′LTR-test-R were used to amplify the region between the 5′ and 3′LTR sequences. The expected size of the amplicons was 609 bp. The amplicon sequences were determined by sequencing analysis and are shown at the bottom. WT genomic DNA isolated from TU wild-type zebrafish, KO genomic DNA isolated from ERV-knockout zebrafish, M DNA ladder. (d) Spinal abnormality was caused by various sgRNAs in ZFERV-knockout (KO) zebrafish embryos. ZFERV knockout embryos produced by separately injecting LTR-sgRNA (a), sgRNA1 (b), sgRNA2 (c), or sgRNA3 (d) into the Cas9-transgenic zebrafish embryos. e Normal spinal morphology in wild-type embryos
To confirm that the abnormality was not caused by off-target effects, three additional sgRNAs target to conserved sequence in 5′LTR and 3′LTR of ZFERV were designed (the sgRNA sequences are shown in Supplemental Table 2) and separately injected into Cas9-transgenic zebrafish embryos. In line with the notion that the spinal abnormality was caused by ZFERV deletion, spinal abnormalities (Fig. 4d) and dramatic reductions in of the Env gene expression (Supplemental Fig. 6) were also observed in embryos treated with these additional sgRNAs. Embryos with spinal abnormalities constituted 84% ~ 89.6% of total embryos (Supplemental Table 3).
To exclude possible toxic effects of constitutive transgenic Cas9 expression on spinal development, fertilized wild-type zebrafish eggs were injected with Cas9 mRNA and sgRNAs targeted to ERV. As expected, embryos that developed from these eggs also showed spinal abnormalities, though the abnormal embryos constituted only approximately 30% of total embryos. Correspondingly, Env gene expression in the abnormal embryos was reduced by approximately 60% compared with wild-type embryos (Fig. 5a).
Quantitative analysis of Env, Delta D, and notch1 gene expression. a Env gene expression in TU wild-type zebrafish and ERV-knockout zebrafish generated from Cas9 transgenic fish (KO1) or by microinjection of Cas9 mRNA and LTR sgRNAs into fertilized eggs (KO2). The expression level is normalized to that in TU wild-type zebrafish. ***p < 0.001. b Delta D and Notch1 gene expression in TU wild-type zebrafish and ERV-knockout zebrafish generated from Cas9-transgenic fish (KO1) or by microinjection of Cas9 mRNA and LTR sgRNAs into fertilized eggs (KO2). The expression level is normalized to that in TU wild-type zebrafish (Values are adjusted mean ± SEM, n = 3 for WT, n = 5 for ZFERV KO). ***p < 0.001
Delta/notch signaling may mediate the role of ERV in spine development
To understand how ERV deletion causes spine abnormality, quantitative PCR analysis was performed to assess the expression of the Delta D and Notch1 genes using RNA isolated from ERV-deficient zebrafish with abnormal spines. Delta D and Notch1 expression was significantly lower in zebrafish with abnormal spines than that in wild-type zebrafish (Fig. 5b), suggesting that ZFERV may be associated with spinal development and that the Delta/Notch signaling pathway may mediate the role of ZFERV. The detection of Her1, which is associated with spinal development in the notch1 signaling pathway is also shown in Supplemental Fig. 7. Her1 expression was significantly reduced in ZFERV knockout samples.
Conclusion
In summary, a Cas9 transgenic zebrafish line was successfully established and was employed in establishing an ERV-deficient zebrafish model. Spinal abnormality is a major phenotype in ERV-deficient zebrafish, which is reported for the first time in this study. This phenotype may be attributed to the suppression of Delta D and Notch 1 gene expression due to ZFERV deletion, suggesting that the role of the ERV in spinal development may be mediated by Delta/Notch signaling.
Abbreviations
- Cas9:
-
CRISPR-associated protein 9
- CRISPR:
-
Clustered Regularly Interspaced Short Palindromic Repeats
- Eef1g:
-
Eukaryotic translation elongation factor 1 gamma
- Ef1α:
-
Elongation factor 1
- eGFP:
-
Enhanced green fluorescent protein
- Env:
-
Envelop
- ERV:
-
Endogenous retrovirus
- HR:
-
Homologous recombination arm
- LTR:
-
Long terminal repeat
- Mitfα:
-
Microphthalmia-associated transcription factor α
- NLS:
-
Nuclear localization signal
- RT-PCR:
-
Reverse transcription PCR
- sgRNA:
-
Single guide RNA
- Tyr:
-
Tyrosinase
- ZFERV:
-
Zebrafish endogenous retrovirus
References
Burket CT, Montgomery JE, Thummel R, Kassen SC, LaFave MC et al (2008) Generation and characterization of transgenic zebrafish lines using different ubiquitous promoters. Transgenic Res 17:265–279
Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W et al (2013) Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell 155:1479–1491
Chen S, Lee B, Lee AY, Modzelewski AJ, He L (2016) Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes. J Biol Chem 291:14457–14467
Chuong EB, Elde NC, Feschotte C (2016) Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351:1083–1087
Jao LE, Wente SR, Chen W (2013) Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci USA 110:13904–13909
Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS (2009) Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res 19:1279–1288
Kraft K, Geuer S, Will AJ, Chan WL, Paliou C et al (2015) Deletions, inversions, duplications: engineering of structural variants using CRISPR/Cas in mice. Cell Reports 10:833–839
Labun K, Montague TG, Gagnon JA, Thyme SB, Valen E (2016) CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res 44:W272–276
Lin S, Staahl BT, Alla RK, Doudna JA (2014) Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. Elife 3:e04766
Liu WY, Zhang KJ (2011) Rapid salt-extraction of genomic DNA from formalin-fixed toad and frog tissues for PCR-based analyses. Asian J Anim Vet Adv 6:958–965
Moon HY, Kim OH, Kim HT, Choi JH, Yeo SY et al (2013) Establishment of a transgenic zebrafish EF1 alpha: Kaede for monitoring cell proliferation during regeneration. Fish Shellfish Immunol 34:1390–1394
Rosen JN, Sweeney MF, Mably JD (2009) Microinjection of zebrafish embryos to analyze gene function. J Vis Exp. https://doi.org/10.3791/1115
Shi J, Zhang H, Gong R, Xiao G (2015) Characterization of the fusion core in zebrafish endogenous retroviral envelope protein. Biochem Biophys Res Commun 460:633–638
Varshney GK, Pei W, LaFave MC, Idol J, Xu L et al (2015) High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res 25:1030–1042
Wen KJ, Yang LJ, Xiong TL, Di C, Ma DH et al (2016) Critical roles of long noncoding RNAs in Drosophila spermatogenesis. Genome Res 26:1233–1244
Acknowledgments
This work was supported by the National Natural Science Foundation of China (91540117, 81773013), the National Key Research and Development Program in China (2016YFC1303604), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (Animal Science and Veterinary Medicine).
Supporting information
Supplemental Table 1—Oligoes used in this article. Supplemental Table 2—The sgRNAs used in generation of ZFERV-deficient zebrafish. Supplemental Table 3—The percentages of abnormal embryos resulted from injection of different sgRNAs into Cas9 transgenic zebrafish embryos. Supplemental Fig. 1—Map of pcDNA3.1[CMV-eGFP] plasmid. Supplemental Fig. 2—Map of pcDNA3.1[eGFP] plasmid. Supplemental Fig. 3—Map of pcDNA3.1[ef1a-eGFP] plasmid. Supplemental Fig. 4—Map of pcDNA3.1[eef1g-eGFP] plasmid. Supplemental Fig. 5—Diagram illustrates location of the primers used to amplify 5′-LTR sequence using genomic DNA from ZFERV knockout (KO) or wild type (WT) zebrafish. An amplicon (266 bp in size) can be produced using the primers and the genomic DNA from WT zebrafish, while no amplicons can be synthesized using the primers and the genomic DNA from ZFERV KO zebrafish. The blue box indicates the 5′-LTR sequence of ZFERV. Supplemental Fig. 6—The expression of Env gene in ZFERV knockout (KO) zebrafish embryos (KO1/KO2/KO3) resulted from injection of different sgRNAs (sgRNA1/sgRNA2/ sgRNA3) into the Cas9 transgenic zebrafish embryos vs. that in wild type (WT) embryos. The expression of Env gene was determined by quantitative PCR with Env-specific primers. The expression of Env gene was expressed as fold change over that in WT embryos. The internal reference genes include beta-actin and GAPDH. Data are presented as mean ± SEM. n = 3 for WT, n = 5 for each ZFERV-KO group. *** denotes statistical significance (p < 0.001) against WT embryos. Supplemental Fig. 7—The expression of Her1 gene in ZFERV knockout (KO) zebrafish and wildtype control (WT). The expression of Her1 gene was determined by quantitative PCR with Her1-specific primers. The expression of Her1 gene was expressed as fold change over that in WT embryos. The internal reference genes include beta-actin and GAPDH. Data are presented as mean ± SEM. n= 3 for WT, n = 5 for each ZFERV-KO group. *** denotes statistical significance (p < 0.001) against WT embryos.
Author information
Authors and Affiliations
Contributions
This study was conceived by H. C. and designed by H.C. and Z.Y. The experiments were conducted by Z. Y., S. C., S. X., X. L., Z. S., Y. Y., and X.H. The manuscript was drafted by T.G. and Z. Y. All authors read and provided constructive comments on the manuscript. H. C. finalized the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Yang, Z., Chen, S., Xue, S. et al. Generation of Cas9 transgenic zebrafish and their application in establishing an ERV-deficient animal model. Biotechnol Lett 40, 1507–1518 (2018). https://doi.org/10.1007/s10529-018-2605-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-018-2605-5