Abstract
Objectives
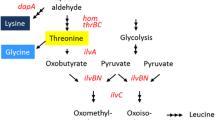
To explore systemic effects of mutations in the UDP-N-acetylmuramoylalanyl-d-glutamate 2,6-diaminopimelate ligase (MurE) of Corynebacterium glutamicum, that leads to extracellular l-lysine accumulation by this bacterium.
Results
The analysis of a mutant cohort of C. glutamicum strains carrying all possible 20 amino acids at position 81 of MurE revealed unexpected effects on cellular properties. With increasing l-lysine accumulation the growth rate of the producing strain is reduced. A dynamic flux balance analysis including the flux over MurE fully supports this finding and suggests that further reductions at this flux control point would enhance l-lysine accumulation even further. The strain carrying the best MurE variant MurE-G81K produces 37 mM l-lysine with a yield of 0.17 g/g (l-lysine·HCl/glucose·H2O), bearing no other genetic modification. Interestingly, among the strains with high l-lysine titers, strain variants occur which, despite possessing the desired amino acid substitutions in MurE, have regained close to normal growth and correspondingly lower l-lysine accumulation. Genome analyses of such variants revealed the transposition of mobile genetic elements which apparently annulled the favorable consequences of the MurE mutations on l-lysine formation.
Conclusion
MurE is an attractive target to achieve high l-lysine accumulation, and product formation is inversely related to the specific growth rate. Moreover, single point mutations leading to elevated l-lysine titers may cause systemic effects on different levels comprising also major genome modifications. The latter caused by the activity of mobile genetic elements, most likely due to the stress conditions being characteristic for microbial metabolite producers.



Similar content being viewed by others
References
Basavannacharya C, Moody PR, Munshi T, Cronin N, Keep NH, Bhakta S (2010) Essential residues for the enzyme activity of ATP-dependent mure ligase from Mycobacterium tuberculosis. Protein Cell 1:1011
Becker J, Zelder O, Häfner S, Schröder H, Wittmann C (2011) From zero to hero—design-based systems metabolic engineering of Corynebacterium glutamicum for l-lysine production. Metab Eng 13:159–168
Binder S, Schendzielorz G, Stäbler N, Krumbach K, Hoffmann K, Bott M, Eggeling L (2012) A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol 13:R40
Binder S, Siedler S, Marienhagen J, Bott M, Eggeling L (2013) Recombineering in Corynebacterium glutamicum combined with optical nanosensors: a general strategy for fast producer strain generation. Nucleic Acids Res 41:6360–6369
Eggeling L, Bott M (2015) A giant market and a powerful metabolism: l-lysine provided by Corynebacterium glutamicum. Appl Microbiol Biotechnol 99:3387–3394
Gomez J, Höffner K, Barton P (2014) DFBAlab: a fast and reliable matlab code for dynamic flux balance analysis. BMC Bioinform 15:1–10
Kalinowski J (2005) The genomes of amino acid-producing Corynebacteria. In: Eggeling, Bott (eds) Handbook of Corynebacterium glutamicum. Taylor & Francis, Boca Raton, pp 37–56
Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, Dusch N, Eggeling L, Eikmanns BJ, Gaigalat L, Goesmann A, Hartmann M, Huthmacher K, Krämer R, Linke B, McHardy AC, Meyer F, Möckel B, Pfefferle W, Pühler A, Rey DA, Rückert C, Rupp O, Sahm H, Wendisch VF, Wiegräbe I, Tauch A (2003) The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol 104:5–25
Lee JH, Sung BH, Kim MS, Blattner FR, Yoon BH, Kim JH, Kim SC (2009) Metabolic engineering of a reduced-genome strain of Escherichia coli for l-threonine production. Microb Cell Fact 8:2
Mengin-Lecreulx D, Parquet C, Desviat LR, Plá J, Flouret B, Ayala JA, van Heijenoort J (1989) Organization of the murE–murG region of Escherichia coli: identification of the murD gene encoding the d-glutamic-acid-adding enzyme. J Bacteriol 171:6126–6134
Möker N, Brocker M, Schaffer S, Krämer R, Morbach S, Bott M (2004) Deletion of the genes encoding the MtrA–MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol Microbiol 54:420–438
Pósfai G, Plunkett G 3rd, Fehér T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, Burland V, Harcum SW, Blattner FR (2006) Emergent properties of reduced-genome Escherichia coli. Science 312:1044–1046
Reyes O, Eggeling L (2005) Experiments. In: Eggeling, Bott (eds) Handbook of Corynebacterium glutamicum. Taylor & Francis, Boca Raton, pp 535–566
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73
Söding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248
Zelle E, Nöh K, Wiechert W (2015) Growth and production capabilities of Corynebacterium glutamicum: interrogating a genome-scale metabolic network model. In: Burkowski A (ed) Corynebacterium glutamicum: from systems biology to biotechnological applications. Caister Academic Press, Erlangen, pp 39–54
Acknowledgments
JH was supported by a fellowship from the CLIB Graduate Cluster Industrial Biotechnology (http://www.graduatecluster.net/).
Supporting information
Supplementary procedures—allelic exchange of murE mutations and dynamic flux balance analysis.
Supplementary Table 1—List of plasmids and primers.
Supplementary Table 2—Lysine accumulations and growth rates of clones.
Supplementary Table 3—Structural variants detected by genome sequencing.
Supplementary Fig. 1—Structure of MurE of C. glutamicum.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hochheim, J., Kranz, A., Krumbach, K. et al. Mutations in MurE, the essential UDP-N-acetylmuramoylalanyl-d-glutamate 2,6-diaminopimelate ligase of Corynebacterium glutamicum: effect on l-lysine formation and analysis of systemic consequences. Biotechnol Lett 39, 283–288 (2017). https://doi.org/10.1007/s10529-016-2243-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2243-8