Abstract
Objectives
To confirm the reductive dehalogenation ability of the aerobic strain of Delftia sp. EOB-17, finding more evidences to support the hypothesis that reductive dehalogenation may occur extensively in aerobic bacteria.
Results
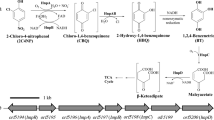
Delftia sp. EOB-17, isolated from terrestrial soil contaminated with halogenated aromatic compounds, completely degraded 0.2 mM DBHB in 28 h and released two equivalents of bromides under aerobic conditions in the presence of sodium succinate. LC–MS analysis revealed that DBHB was transformed to 4-hydroxybenzoate via 3-bromo-4-hydroxybenzoate by successive reductive dehalogenation. Highly conserved DBHB-degrading genes, including reductive dehalogenase gene (bhbA3) and the extra-cytoplasmic binding receptor gene (bhbB3), were also found in strain EOB-17 by genome sequencing. The optimal temperature and pH for DBHB reductive dehalogenation activity are 30 °C and 8, respectively, and 0.1 mM Cd2+, Cu2+, Hg2+ and Zn2+ strongly inhibited dehalogenation activity.
Conclusions
The aerobic strain of Delftia sp. EOB-17 was confirmed to reductively dehalogenate DBHB under aerobic conditions, providing another evidence to support the hypothesis that reductive dehalogenation occurs extensively in aerobic bacteria.





Similar content being viewed by others

References
Adriaens P, Gruden C, McCormick ML (2004) Biogeochemistry of halogenated hydrocarbons. Biogeochemistry 9:A9000
Bergmann JG, Sainik J (1957) Determination of trace amounts of chlorine in naphtha. Anal Chem 29:241–243
Chen K, Huang LL, Xu CF, Liu XM, He J, Zinder SH, Li SP, Jiang JD (2013) Molecular characterization of the enzymes involved in the degradation of a brominated aromatic herbicide. Mol Microbiol 89:1121–1139
de Jong RM, Dijkstra BW (2003) Structure and mechanism of bacterial dehalogenases: different ways to cleave a carbon-halogen bond. Curr Opin Struct Biol 13:722–730
Egland PG, Gibson J, Harwood CS (2001) Reductive, coenzyme A-mediated pathway for 3-chlorobenzoate degradation in the phototrophic bacterium Rhodopseudomonas palustris. Appl Environ Microbiol 67:1396–1399
Gan HM, Chew TH, Tay YL, Lye SF, Yahya A (2012) Genome sequence of Hydrogenophaga sp. strain PBC, a 4-aminobenzenesulfonate-degrading bacterium. J Bacteriol 194:4759–4760
Janssen DB, Oppentocht JE, Poelarends GJ (2001) Microbial dehalogenation. Curr Opin Biotechnol 12:254–258
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306
Kuntze K, Kiefer P, Baumann S, Seifert J, Bergen MV, Vorholt JA, Boll M (2011) Enzymes involved in the anaerobic degradation of meta-substituted halobenzoates. Mol Microbiol 82:758–769
McTamney PM, Rokita SE (2009) A mammalian reductive deiodinase has broad power to dehalogenate chlorinated and brominated substrates. J Am Chem Soc 131:14212–14213
Miyauchi K, Suh SK, Nagata Y, Takagi M (1998) Cloning and sequencing of a 2,5-dichlorohydroquinone reductive dehalogenase gene whose product is involved in degradation of γ-hexachlorocyclohexane by Sphingomonas paucimobilis. J Bacteriol 180:1354–1359
Payne KA, Quezada CP, Fisher K et al (2015) Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation. Nature 517:513–516
Romanov V, Hausinger RP (1996) NADPH-dependent reductive ortho dehalogenation of 2,4-dichlorobenzoic acid in Corynebacterium sepedonicum KZ-4 and coryneform bacterium strain NTB-1 via 2,4-dichlorobenzoyl coenzyme A. J Bacteriol 178:2656–2661
Wen A, Fegan M, Hayward C, Chakraborty S et al (1999) Phylogenetic relationships among members of the Comamonadaceae, and description of Delftia acidovorans (den Dooren de Jong 1926, and Tamaoka et al. 1987) gen. nov., comb. nov. Int J Syst Bacteriol 49:567–576
Willems A, De Vos P (2006) Comamonas. Prokaryotes. Springer, New York, pp 723–736
Xun LY, Topp E, Orserl CS (1992) Purification and characterization of a tetrachloro-p-hydroquinone reductive dehalogenase from a Flavobacterium sp. J Bacteriol 174:8003–8007
Acknowledgments
This work was financially supported by the Chinese National Science Foundation for Excellent Young Scholars (31222003), the Outstanding Youth Foundation of Jiangsu Province (BK20130029), the Program for New Century Excellent Talents in University (NCET-12-0892), the Fundamental Research Funds for the Central Universities (KYZ201422), the National Natural Science Foundation of China (31400105) and the China Postdoctoral Science Foundation (2014M561666).
Supporting information
Supplementary Table 1—Sequence comparison of deduced DBHB-degrading genes in Delftia sp. EOB-17.
Supplementary Fig. 1—Transmission electron micrograph of a negatively stained cell of strain EOB-17 shows a rod (0.7–0.9 μm × 1.7–2.2 μm) shape with polar flagella.
Supplementary Fig. 2—Neighbor-joining phylogenetic tree based on 16S rRNA gene sequences shows the relationship of strain EOB-17 and its related taxa. Bootstrap values (expressed as percentages of 1000 replications) >50 % are shown at branching points. Bar 0.005 substitutions per nucleotide position.
Supplementary Materials and methods.
Supplementary References.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, K., Jian, S., Huang, L. et al. Reductive dehalogenation of 3,5-dibromo-4-hydroxybenzoate by an aerobic strain of Delftia sp. EOB-17. Biotechnol Lett 37, 2395–2401 (2015). https://doi.org/10.1007/s10529-015-1932-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-015-1932-z