Abstract
Previous studies found that the circadian clock gene participated in the genesis and development of breast cancer. However, research findings on the relationship between polymorphisms in the CLOCK gene and breast cancer risk were inconsistent. This study performed a meta-analysis of the association between CLOCK gene polymorphisms and breast cancer risk. PubMed, Cochrane Library, and Embase databases were electronically searched to collect studies on the association between CLOCK gene polymorphisms and breast cancer risk from inception to February 14, 2022. The quality of the included literature was assessed using the Newcastle–Ottawa Scale. For statistical analysis, odds ratio (OR) and 95% confidence intervals (CIs) were calculated using STATA 14.0. In addition, publication bias was performed by the funnel diagram and the Harbord’s regression test. And sensitivity analysis was assessed by the trim and fill method. A total of 6 eligible studies, including 10,164 subjects (5488 breast cancer cases and 4676 controls), were screened in this meta-analysis. Though we did not find a significant association between the polymorphisms in the overall CLOCK gene with breast cancer risk [OR (95%CI) = 0.98 (0.96, 1.01), P = 0.148], we found that compared with T/T types of rs3749474 in CLOCK, T/C and C/C types of rs3749474 were associated with lower risk of breast cancer [OR (95%CI) = 0.93 (0.88, 0.98), P = 0.003]. The sensitivity analysis confirmed the robustness of the results. The funnel plot showed no significant publication bias. Polymorphisms in the CLOCK gene might be associated with breast cancer risk. More studies are needed to confirm the conclusion.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is one of the most common malignancies in women worldwide, and its incidence is increasing yearly (Nounu et al. 2022). It was reported that there were 281,550 diagnosed cases of breast cancer around the world in 2021, and breast cancer was estimated to cause nearly 40,000 deaths, which accounted for 7% of all cancer mortality each year (Ropri et al. 2021). The traditional risk factors of breast cancer include reproductive cycle, lifestyle, obesity, genetic susceptibility, DNA methylation, miRNA, and family history (Cheng et al. 2022; Jiralerspong et al. 2009; Joo et al. 2018; Li et al. 2010; Lin et al. 2022; Wang et al. 2022; Wen et al. 2022; Zhou et al. 2014). These days, large numbers of epidemiological studies have shown that circadian rhythm disruption, which is considered carcinogenic to humans (group 2A), contributes to the increased incidence of breast cancer (Dieterich et al. 2014; Samuelsson et al. 2018). Using data from the Nurses’ Health Study with 0.19 million participants, Wegrzyn et al. found that 20 years or more of night-shift work was associated with a significantly higher risk of breast cancer compared with non-night shift workers [HR(95%CI) = 2.15 (1.23, 2.73)], highlighting the importance of circadian rhythm in the pathogenesis of breast cancer (Wegrzyn et al. 2017).
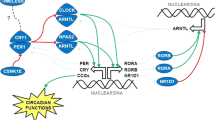
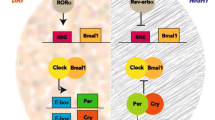
The circadian rhythm is generated and controlled by a series of circadian genes involved in maintaining the internal coordination of multiple oscillators within and between various organ systems to provide the most efficient response to the day/night cycle (Bass and Takahashi 2010). These genes include basic helix-loop-helix ARNT like 1 (BMAL1), Circadian Locomotor Output Cycles Kaput (CLOCK), cryptochrome circadian regulator 1/2, period circadian regulator 1/2/3. (Masri and Sassone-Corsi 2018). Among them, the CLOCK gene was discovered as the first mammalian circadian gene, located in human chromosome 4q12; it codifies the CLOCK protein, a positive regulatory arm of the circadian system (Pagliai et al. 2019). CLOCK plays a role in regulating cell physiological processes, such as the cell cycle, DNA damage response, cell proliferation, and apoptosis. Most epidemiology studies have indicated that CLOCK gene variations are associated with the risk of obesity, cardiovascular diseases, type 2 diabetes, and different types of cancer (Cuninkova and Brown 2008; Valenzuela et al. 2016). However, studies on the effect of CLOCK polymorphisms on breast cancer yield inconsistent results. Zienolddiny et al. found that TT carriers in CLOCK rs3749474 had a reduced risk of breast cancer [OR (95%CI) = 0.64 (0.45, 0.92)] among 563 breast cancer cases and 619 controls (Zienolddiny et al. 2013), while Hoffman et al. did not find the significant association between CLOCK rs3749474 and breast cancer risk (Hoffman et al. 2010). Therefore, the need for further systematical research on the effects of CLOCK polymorphisms on breast cancer remains.
In this study, we conducted a meta-analysis to systematically evaluate the associations between CLOCK gene polymorphisms and breast cancer risk.
Methods
Literature Search Strategy
We prepared this report in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) principle (Liberati et al. 2009). Two researchers, Lixing Wu, and Xuenian Ji, independently searched PubMed, Cochrane Library, and Embase databases, and the search period was set from inception through February 14, 2022. The literature search was conducted according to the search characteristics of each database, and the following keywords were used: ("sleep–wake circadian rhythms" OR "circadian sleep disorders" OR "sleep–wake pattern" OR" sleep disorders" OR "circadian rhythm2 disorder" OR "circadian rhythm") AND ("clock gene" OR "CLOCK") AND ("breast cancer" OR "breast tumor" OR "breast carcinoma"). We first screened the titles and abstracts, then read the full text of all potentially eligible studies. The references of the included literature were also manually searched and reviewed for additional literature.
Inclusion and Exclusion Criteria
Inclusion criteria were as follows: (1) patients definitively diagnosed with breast cancer; (2) definitive detection of CLOCK genotype and allele frequency that were provided in the study; (3) relationships between CLOCK gene polymorphism and the risk of breast cancer that were evaluated; (4) odds ratio (OR), hazard ratio and 95% confidence intervals that could be obtained directly or indirectly calculated based on the data provided in the graphics or tables; (5) study in English.
Exclusion criteria were as follows: (1) duplicate publications or incomplete data; (2) reviews, meta-analysis, only abstracts, conference proceedings, expert reviews, and case reports; (3) literature with unclear diagnostic criteria of breast cancer and genotyping methods; (4) control group with unclear population or history of cervical cancer-related diseases; (5) cell or animal studies.
Data Extraction
Two investigators independently screened the literature and extracted the required information from the literature. The required information included: first author, year of publication, geographic region, CLOCK polymorphisms, sample size, clinical stage, detection method, age, and the number of subjects.
Quality Evaluation
Two researchers independently evaluated the quality of the included literature according to the Newcastle–Ottawa Scale (NOS) criteria and the total score ranging from 0 to 9 (Stang 2010). Studies with a total NOS score ≥ 6 are considered high-quality studies (Xiong et al. 2018).
Statistical Analysis
In this meta-analysis, the pooled odds ratio (OR) with its corresponding 95% confidence intervals (CIs) were used to assess the effect size for studies that reported the association between CLOCK gene polymorphism and the risk of breast cancer. The dominant model, the recessive model, and the over-dominant model were used in the meta-analysis. We used the Q test and I2 statistic to evaluate the heterogeneity across studies. If P ≥ 0.1 and I2 < 50%, the results indicated that the homogeneity among studies was good, and the fixed-effect model was chosen for the meta-analysis; if P < 0.1 and I2 ≥ 50%, the results indicated that there was heterogeneity among studies, and the random-effect model was chosen. Sensitivity analysis was used to evaluate the effect of individual studies on the final results and to determine the reliability of the results. Funnel plots and Harbord’s test was used to assess whether there was significant publication bias. Meta-analyses were performed using STATA 14.0 (Stata Corporation, College Station, Texas, USA). Except for the heterogeneity test, two-sided P values < 0.05 were considered statistically significant.
Results
Literature Search
According to the search strategy, a total of 122 studies were obtained from various databases. Among them, 13 studies were excluded due to duplication; 34 studies were removed due to being reviews, only abstracts, conference literature, or systematic evaluation; 15 animal experiments were excluded; 19 studies with non-specified genes were excluded. Therefore, seven studies were obtained provisionally. In addition, two studies were obtained from reference list searching. After excluding three studies with incomplete data, six studies comprising 10,164 subjects, including 5488 breast cancer cases and 4676 controls, remained in the meta-analysis. The literature search process is detailed in Fig. 1. All 6 studies were case–control studies, and the genotypes of CLOCK were determined by Polymerase Chain Reaction (PCR). Characteristics of the included 6 studies are shown in Table 1. In addition, the NOS quality scores of these studies ranged from 6 to 8 points (Table 2).
Relationship Between CLOCK Polymorphisms and Breast Cancer Risk
A total of 13 CLOCK gene SNPs were included. The dominant model, over-dominant model, and recessive model were used in the meta-analysis. However, we did not find a significant association between CLOCK polymorphisms and breast cancer [dominant model: OR (95%CI) = 0.98 (0.91, 1.04), Fig. 2; over-dominant model: OR (95%CI) = 1.00 (0.96, 1.05), Fig. 3; recessive model: OR (95%CI) = 0.98 (0.94, 1.03), Fig. 4].
A total of three studies examined the association between rs3749474 and breast cancer risk. In the sub-meta-analysis of rs3749474, we found that compared with T/T types of rs3749474, T/C and C/C types of rs3749474 were associated with lower risk of breast cancer [OR (95%CI) = 0.91 (0.85, 0.97), dominant model], with non-significant heterogeneity across studies (I2 < 0.001 and Heterogeneity = 0.971). The results of the meta-analysis are shown in Fig. 5.
Publication Bias
The Cochrane Collaboration’s tool for assessing the risk of bias is shown in Fig. 6. As expected, only the two full-paper trials were of high quality. The funnel diagram is shown in Fig. 7. Harbord's regression test (P = 0.864) indicated no publication bias.
Sensitivity Analysis
The leave-one-out sensitivity analysis showed that no single study significantly affected the pooled correlation from the meta-analysis (the OR ranging from 0.98 to 1.24), which indicated the reliability of the findings. The trim and fill method suggests funnel plot symmetry (Fig. 8).
Discussion
To provide insight into the relationship between CLOCK polymorphisms and breast cancer risk, we performed a meta-analysis among 10,164 participants, including 5488 breast cancer cases and 4676 controls. We found that the mutation types of one CLOCK SNP, rs3749474 were significantly associated with the risk of breast cancer. Compared with T/T types of rs3749474, T/C and C/C types of rs3749474 were associated with a lower risk of breast cancer [OR (95%CI) = 0.93 (0.88, 0.98)]. These results uncover the important role of CLOCK polymorphisms in breast cancer.
Breast cancer is one of the most common cancer types among women. Further understanding of progress, treatments, and biomarkers of breast cancer is urgently needed. A number of risk factors have been found to be associated with the risk of breast cancer these years, including cytokine, miRNA, and trace elements in food, etc. (Çetin and Topçul 2022; Cheng et al. 2022; Majed 2022; Wang et al. 2022; Xin and Zhiyuan 2022; Zhou et al. 2022). Based on these, molecular markers and reference laboratory tests for breast cancer diagnosis and prognosis have been developed, but the methods are limited to specific subtypes (Park et al. 2017). Thus, we still need novel approaches to the diagnosis of breast cancer.
It has been reported that disruption in sleep and circadian rhythm disorders are significantly associated with the risk of breast cancer (He et al. 2015). An animal study by Zhang et al. showed that disturbances of the circadian system through ablation of the pineal gland or constant light exposure could result in breast carcinogenesis (Zhang et al. 2020). In humans, a meta-analysis of 26 studies with more than one million participants found that female flight attendants who worked long shifts at night had an increased incidence of breast cancer (Manouchehri et al. 2021). The mechanisms may lie in the secretion disorder of nocturnal melatonin in the pineal gland, the activities of the hormone estrogen and estrogen receptor, and the circadian clock genes (Stevens 2005; Stevens and Davis 1996; Stevens and Rea 2001).
Evidence has also shown that individual genes of the circadian clock play a role in controlling tumorigenesis. CLOCK is one of the core circadian clock genes. Its corresponding protein belongs to the basic helix-loop-helix PAS family of transcription factors and forms heterodimers with BMAL1 to enhance target gene expression (Benna et al. 2017). CLOCK has been identified as a significant modifier of breast cancer incidence (Sancar and Gelder 2021). On the one hand, the expression of oncogene c-Myc could be controlled by CLOCK; on the other hand, oncogenes c-Myc, P53, and Ras could affect the face of CLOCK (Sancar and Gelder 2021). Moreover, CLOCK could induce remodeling of the tumor microenvironment cells by disturbing the cellular metabolism, altering gene expression, and aberrantly activating signaling pathways (Malla et al. 2021). Using state-of-the-art immune cell deconvolution and pathway quantification, Wu et al. demonstrated that abnormal expression of CLOCK contributed to T cell exhaustion and global upregulation of immune inhibitory molecules (Wu et al. 2019).
SNP rs3749474 is located in the CLOCK 3’-untranslated region. In the current study, we performed the sub-group meta-analysis only for rs3749474, and we found that compared with wild type T/T, the homozygous mutant-type C/C and heterozygous-type T/C of rs3749474 were associated with a lower risk of breast cancer. However, rs3749474 is not specific to breast cancer. Zhou et al. found that the homozygous mutant-type of rs3749474 was associated with better survival of colorectal cancer (F. Zhou et al. 2012). It has also been reported that the rs3749474 polymorphism could modulate the effect of energy intake on nutritional status (Camblor Murube et al. 2020; Espinosa-Salinas et al. 2020). The biological role of rs3749474 on breast cancer still needs further investigation.
To our knowledge, this is the first meta-analysis of the association between CLOCK polymorphisms and breast cancer risk, which incorporates multiple SNPs. Our findings may gain insights into breast cancer genetics and uncover the roles of the circadian clock gene on breast cancer, which helps provide a basis for further research in breast cancer diagnosis and treatment. However, the present study has some limitations. First, we could not conduct a subgroup meta-analysis given the limited data in the included studies, so our results should be interpreted cautiously. Second, only studies published in English were included; therefore, we might have missed some relevant studies in other languages. Third, bias might have been introduced due to confounding factors among different studies.
Conclusion
The mutation types of CLOCK gene rs3749474 are negatively associated with the risk of breast cancer. More studies are still needed to confirm this conclusion.
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Bass J, Takahashi JS (2010) Circadian integration of metabolism and energetics. Science 330:1349–1354. https://doi.org/10.1126/science.1195027
Benna C, Helfrich-Förster C, Rajendran S, Monticelli H, Pilati P, Nitti D, Mocellin S (2017) Genetic variation of clock genes and cancer risk: a field synopsis and meta-analysis. Oncotarget 8:23978–23995. https://doi.org/10.18632/oncotarget.15074
Camblor Murube M, Borregon-Rivilla E, Colmenarejo G, Aguilar-Aguilar E, Martínez JA, Ramírez De Molina A, Reglero G, Loria-Kohen V (2020) Polymorphism of CLOCK gene rs3749474 as a modulator of the circadian evening carbohydrate intake impact on nutritional status in an adult sample. Nutrients. https://doi.org/10.3390/nu12041142
Çetin İ, Topçul M (2022) Investigation of the effects of the endogenous cannabinoid anandamide on luminal a breast cancer cell line MCF-7. Cell Mol Biol (Noisy-le-grand) 68:129–133. https://doi.org/10.14715/cmb/2022.68.4.16
Cheng H, Chen L, Fang Z, Wan Q, Du Z, Ma N, Guo G, Lu W (2022) The effect of miR-138 on the proliferation and apoptosis of breast cancer cells through the NF-κB/VEGF signaling pathway. Cell Mol Biol (Noisy-le-grand) 68:132–137. https://doi.org/10.14715/cmb/2022.68.2.19
Cheng H, Chen L, Fang Z, Wan Q, Du Z, Ma N, Guo G, Lu W (2022) STIM2 promotes the invasion and metastasis of breast cancer cells through the NFAT1/TGF-β1 pathway. Cell Mol Biol (Noisy-le-grand) 67:55–61. https://doi.org/10.14715/cmb/2021.67.6.8
Cuninkova L, Brown SA (2008) Peripheral circadian oscillators: interesting mechanisms and powerful tools. Ann NY Acad Sci 1129:358–370. https://doi.org/10.1196/annals.1417.005
Dieterich M, Stubert J, Reimer T, Erickson N, Berling A (2014) Influence of lifestyle factors on breast cancer risk. Breast Care (basel) 9:407–414. https://doi.org/10.1159/000369571
Espinosa-Salinas I, San-Cristobal R, Colmenarejo G, Loria-Kohen V, Molina S, Reglero G, Ramirez de Molina A, Martinez JA (2020) Polymorphic appetite effects on waist circumference depend on rs3749474 CLOCK gene variant. Nutrients. https://doi.org/10.3390/nu12061846
He C, Anand ST, Ebell MH, Vena JE, Robb SW (2015) Circadian disrupting exposures and breast cancer risk: a meta-analysis. Int Arch Occup Environ Health 88:533–547. https://doi.org/10.1007/s00420-014-0986-x
Hoffman AE, Yi CH, Zheng T, Stevens RG, Leaderer D, Zhang Y, Holford TR, Hansen J, Paulson J, Zhu Y (2010) CLOCK in breast tumorigenesis: genetic, epigenetic, and transcriptional profiling analyses. Cancer Res 70:1459–1468. https://doi.org/10.1158/0008-5472.CAN-09-3798
Jiralerspong S, Palla SL, Giordano SH, Meric-Bernstam F, Liedtke C, Barnett CM, Hsu L, Hung MC, Hortobagyi GN, Gonzalez-Angulo AM (2009) Metformin and pathologic complete responses to neoadjuvant chemotherapy in diabetic patients with breast cancer. J Clin Oncol 27:3297–3302. https://doi.org/10.1200/jco.2009.19.6410
Joo JE, Dowty JG, Milne RL, Wong EM, Dugué PA, English D, Hopper JL, Goldgar DE, Giles GG, Southey MC (2018) Heritable DNA methylation marks associated with susceptibility to breast cancer. Nat Commun 9:867. https://doi.org/10.1038/s41467-018-03058-6
Li CI, Chlebowski RT, Freiberg M, Johnson KC, Kuller L, Lane D, Lessin L, O’Sullivan MJ, Wactawski-Wende J, Yasmeen S, Prentice R (2010) Alcohol consumption and risk of postmenopausal breast cancer by subtype: the women’s health initiative observational study. J Natl Cancer Inst 102:1422–1431. https://doi.org/10.1093/jnci/djq316
Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D (2009) The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ 339:B2700. https://doi.org/10.1136/bmj.b2700
Lin J, Ding Q, Zhang G, Yin X (2022) Study on PI3k gene expression in breast cancer samples and its association with clinical factors and patient survival. Cell Mol Biol (Noisy-le-grand) 67:321–327. https://doi.org/10.14715/cmb/2021.67.4.36
Majed SO (2022) RNA sequencing-based total RNA profiling; the oncogenic MiR-191 identification as a novel biomarker for breast cancer. Cell Mol Biol (Noisy-le-grand) 68:177–191. https://doi.org/10.14715/cmb/2022.68.1.22
Malla RR, Padmaraju V, Amajala KC, Chalikonda G, Nagaraju GP (2021) Association between the Circadian clock and the tumor microenvironment in breast cancer. Crit Rev Oncog 26:43–51. https://doi.org/10.1615/CritRevOncog.2021040504
Manouchehri E, Taghipour A, Ghavami V, Ebadi A, Homaei F, Latifnejad Roudsari R (2021) Night-shift work duration and breast cancer risk: an updated systematic review and meta-analysis. BMC Womens Health 21:89. https://doi.org/10.1186/s12905-021-01233-4
Masri S, Sassone-Corsi P (2018) The emerging link between cancer, metabolism, and circadian rhythms. Nat Med 24:1795–1803. https://doi.org/10.1038/s41591-018-0271-8
Nounu A, Kar SP, Relton CL, Richmond RC (2022) Sex steroid hormones and risk of breast cancer: a two-sample Mendelian randomization study. Breast Cancer Res 24:66. https://doi.org/10.1186/s13058-022-01553-9
Pagliai G, Sofi F, Dinu M, Sticchi E, Vannetti F, Molino Lova R, Ordovas JM, Gori AM, Marcucci R, Giusti B, Macchi C (2019) CLOCK gene polymorphisms and quality of aging in a cohort of nonagenarians—the MUGELLO study. Sci Rep 9:1472. https://doi.org/10.1038/s41598-018-37992-8
Park SM, Choi EY, Bae M, Choi JK, Kim YJ (2017) A long-range interactive DNA methylation marker panel for the promoters of HOXA9 and HOXA10 predicts survival in breast cancer patients. Clin Epigenetics 9:73. https://doi.org/10.1186/s13148-017-0373-z
Ropri AS, DeVaux RS, Eng J, Chittur SV, Herschkowitz JI (2021) Cis-acting super-enhancer lncRNAs as biomarkers to early-stage breast cancer. Breast Cancer Res 23:101. https://doi.org/10.1186/s13058-021-01479-8
Samuelsson LB, Bovbjerg DH, Roecklein KA, Hall MH (2018) Sleep and circadian disruption and incident breast cancer risk: an evidence-based and theoretical review. Neurosci Biobehav Rev 84:35–48. https://doi.org/10.1016/j.neubiorev.2017.10.011
Sancar A, Van Gelder RN (2021) Clocks, cancer, and chronochemotherapy. Science. https://doi.org/10.1126/science.abb0738
Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25:603–605. https://doi.org/10.1007/s10654-010-9491-z
Stevens RG (2005) Circadian disruption and breast cancer: from melatonin to clock genes. Epidemiology 16:254–258. https://doi.org/10.1097/01.ede.0000152525.21924.54
Stevens RG, Davis S (1996) The melatonin hypothesis: electric power and breast cancer. Environ Health Perspect 104(Suppl 1):135–140. https://doi.org/10.1289/ehp.96104s1135
Stevens RG, Rea MS (2001) Light in the built environment: potential role of circadian disruption in endocrine disruption and breast cancer. Cancer Causes Control 12:279–287. https://doi.org/10.1023/a:1011237000609
Valenzuela FJ, Vera J, Venegas C, Munoz S, Oyarce S, Munoz K, Lagunas C (2016) Evidences of polymorphism associated with circadian system and risk of pathologies: a review of the literature. Int J Endocrinol 2016:2746909. https://doi.org/10.1155/2016/2746909
Wang D, Wang C, Sun L, Lu X, Shi J, Chen J, Zhang X (2022) MiR-143–3p increases the radiosensitivity of breast cancer cells through FGF1. Cell Mol Biol (Noisy-le-grand) 67:256–262. https://doi.org/10.14715/cmb/2021.67.5.35
Wegrzyn LR, Tamimi RM, Rosner BA, Brown SB, Stevens RG, Eliassen AH, Laden F, Willett WC, Hankinson SE, Schernhammer ES (2017) Rotating night-shift work and the risk of breast cancer in the nurses’ health studies. Am J Epidemiol 186:532–540. https://doi.org/10.1093/aje/kwx140
Wen R, Lin H, Li X, Lai X, Yang F (2022) The Regulatory mechanism of EpCAM N-Glycosylation-Mediated MAPK and PI3K/Akt pathways on epithelial-mesenchymal transition in breast cancer cells. Cell Mol Biol (Noisy-le-grand) 68:192–201. https://doi.org/10.14715/cmb/2022.68.5.26
Wu Y, Tao B, Zhang T, Fan Y, Mao R (2019) Pan-cancer analysis reveals disrupted circadian clock associates with T cell exhaustion. Front Immunol 10:2451. https://doi.org/10.3389/fimmu.2019.02451
Xin L, Zhiyuan X (2022) Evaluating serum level of granulocyte, macrophage and granulocyte-macrophage colony-stimulating factors in patients with breast tumor. Cell Mol Biol (Noisy-le-grand) 68:146–152. https://doi.org/10.14715/cmb/2022.68.5.20
Xiong H, Yang Y, Yang K, Zhao D, Tang H, Ran X (2018) Loss of the clock gene PER2 is associated with cancer development and altered expression of important tumor-related genes in oral cancer. Int J Oncol 52:279–287. https://doi.org/10.3892/ijo.2017.4180
Zhang M, Lu Y, Chen Y, Zhang Y, Xiong B (2020) Insufficiency of melatonin in follicular fluid is a reversible cause for advanced maternal age-related aneuploidy in oocytes. Redox Biol 28:101327. https://doi.org/10.1016/j.redox.2019.101327
Zhou F, He X, Liu H, Zhu Y, Jin T, Chen C, Qu F, Li Y, Bao G, Chen Z, Xing J (2012) Functional polymorphisms of circadian positive feedback regulation genes and clinical outcome of Chinese patients with resected colorectal cancer. Cancer 118:937–946. https://doi.org/10.1002/cncr.26348
Zhou W, Ding Q, Pan H, Wu N, Liang M, Huang Y, Chen L, Zha X, Liu X, Wang S (2014) Risk of breast cancer and family history of other cancers in first-degree relatives in Chinese women: a case control study. BMC Cancer 14:662. https://doi.org/10.1186/1471-2407-14-662
Zhou T, Zhou M, Tong C, Zhuo M (2022) Cauliflower bioactive compound sulforaphane inhibits breast cancer development by suppressing NF-κB /MMP-9 signaling pathway expression. Cell Mol Biol (Noisy-le-grand) 68:134–143. https://doi.org/10.14715/cmb/2022.68.4.17
Zienolddiny S, Haugen A, Lie JA, Kjuus H, Anmarkrud KH, Kjaerheim K (2013) Analysis of polymorphisms in the circadian-related genes and breast cancer risk in Norwegian nurses working night shifts. Breast Cancer Res 15:R53. https://doi.org/10.1186/bcr3445
Funding
This work was supported by “the Clinical study on the whole process management of TCM appropriate technology for sleep disorders” and “To investigate the mechanism of Long-Bei-Xiao-Yao-San (LBXYS) inhibiting breast cancer’s effect through the demethylation of miR-145 and activating the miR-145/c-Myc/p53 pathway”. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by YS, LW, XJ, YL and ZZ. The first draft of the manuscript was written by YS and all authors commented on previous versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical Approval
Ethical approval and informed consent is not applicable in this study.
Consent to Publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shi, Y., Wu, L., Ji, X. et al. Relationship Between Breast Cancer Risk and Polymorphisms in CLOCK Gene: A Systematic Review and Meta-Analysis. Biochem Genet 61, 2348–2362 (2023). https://doi.org/10.1007/s10528-023-10372-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10372-2