Abstract
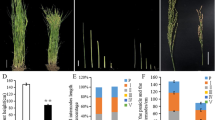
Rice tillering ability and plant height are two of the important traits determining the grain yield. A novel rice (Oryza sativa L.) mutant dhta-34 from an Indica cultivar Zhenong 34 treated by ethyl methy1 sulfonate (EMS) was investigated in this study. The dhta-34 mutant significantly revealed thrifty tillers with reduced plant height, smaller panicles and lighter grains. It also exhibited late-maturing (19.80 days later than the wild type) and withered leaf tip during the mature stage. The length of each internode was reduced compared to the wild type, belonging to the dn type (each internode of the plant stem decreased in the same ratio). The longitudinal section of dhta-34 internodes showed that the length of cells was reduced leading to the dwarfism of the mutant. The F2 population derived from a cross between dhta-34 and an Japonica cultivar Zhenongda 104 were used for gene mapping by using the map-based cloning strategy. The gene DHTA-34 was fine mapped in 183.8kb region flanked by markers 3R-7 and 3R-10. The cloning and sequencing of the target region from the mutant revealed that there was a substitution of G to A in the second exon of LOC_Os03g10620, which resulted in an amino acid substitution arginine to histidine. DHTA-34 encoded a protein of the α/β-fold hydrolase superfamily, which could suppress the tillering ability of rice. DHTA-34 was a strong loss-of-function allele of the Arabidopsis thaliana D14 gene, which was involved in part of strigolactones (SLs) perception and signaling. Moreover, the relative expression of DHTA-34 gene in leaf was higher than that in bud, internode, root or sheath. This study revealed that DHTA-34 played an important role in inhabiting tiller development in rice and further identifying the function of D14.








Similar content being viewed by others
Abbreviations
- dhta-34 :
-
Dwarfism with high tillering ability 34
- EMS:
-
Ethyl methyl sulfonate
- FAA:
-
Formalin-acetic acid-alcohol
- SLs:
-
Strigolactones
References
Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M et al (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51:1019–1029
Arite T, Umehara M, Ishikawa S, Hanada A, Maekawa M, Yamaguchi S et al (2009) D14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol 50(8):1416–1424
Beveridge CA (2000) Long–distance signaling and a mutational analysis of branching in pea. Plant Growth Regul 32:193–203
Beveridge CA, Kyozuka J (2010) New genes in the strigolactone–related shoot branching pathway. Curr Opin Plant Biol 13:34–39
Beveridge CA, Weller JL, Singer SR, Hofer JMI (2003) Axillary meristem development, budding relationships between networks controlling flowering, branching and photoperiod respon siveness. Plant Physiol 131:927–934
Booker J, Auldridge M, Wills S, McCarty D, Klee H, Leyser O (2004) MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule. Curr Biol 14:1232–1238
Booker J, Sieberer T, Wright W, Williamson L, Willett B, Stirnberg P et al (2005) MAX1 encodes a cytochrome P450 family member that acts downstream of MAX3/4 to produce a carotenoid–derived branch–inhibiting hormone. Dev Cell 8:443–449
Braun N, De Saint A, Germain JP, Pillot SBoutet-Mercey, M. Dalmais, I. Antoniadi, C. Rameau, et al (2012) The Pea TCP Transcription Factor PsBRC1 Acts Downstream of Strigolactones to Control Shoot Branching. Plant Physiol 158:225–238
Chevalier F, Nieminen K, Sánchez-Ferrero JC, Rodríguez ML, Chagoyen M, Hardtke CS, Cubas P (2014) Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. Plant Cell 26:1134–1150
Doebley J, Stec A, Gustus C (1995) Teosinte branched 1 and the origin of maize, evidence for epistasis and the evolution of dominance. Genetics 141:333–346
Doebley J, Stec A, Hubbard L (1997) The evolution of apical dominance in maize. Nature 386:485–488
Han L, Mason M, Risseeuw EP, Crosby WL, Somers DE (2004) Formation of an scf ztl complex is required for proper regulation of circadian timing. Plant J 40(2):11
Hubbard L, McSteen P, Doebley J, Hake S (2002) Expression patterns and mutant phenotype of teosinte branched1 correlate with growth suppression in maize and teosinte. Genetics 162:1927–1935
Ishikawa S, Maekawa M, Arite T, Onish K, Takamure I, Kyozuka J (2005) Suppression of tiller bud activity in tillering dwarf mutants of rice. Plant Cell Physiol 46:79–86
Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L et al (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504:401–405
Kang TJ, Yang MS (2004) Rapid and reliable extraction of genomic DNA from various wild–type and transgenic plants. BMC Biotech 4:20–20
Kim HK, Luquet D, Van Oosterom E, Dingkuhn M, Hammer G (2010) Regulation of tillering in sorghum, genotypic effects. Ann Bot 106:69–78
Kulkarni KP, Vishwakarma C, Sahoo SP, Lima JM, Nath M, Dokku P et al (2014) A substitution mutation in OsCCD7 cosegregates with dwarf and increased tillering phenotype in rice. J Genet 93:389–401
Leyser O (2011) Auxin, self–organisation, and the colonial nature of plants. Curr Biol 21:R331–R337
Li X, Qian Q, Fu Z, Wang Y, Xiong G, Zeng D et al (2003) Control of tillering in rice. Nature 422:618–621
Liang WH, Shang F, Lin QT, Lou C, Zhang J (2014) Tillering and panicle branching genes in rice. Gene 537:1–5
Lin H, Wang R, Qian Q, Yan M, Meng X, Fu Z et al (2009) DWARF27, an iron–containing protein required for the biosynthesis of strigolactones, regulates rice tiller bud outgrowth. Plant Cell 21:1512–1525
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the \(2^{-\Delta\Delta C_t}\) method. Methods 25:402–408
McSteen P (2009) Hormonal regulation of branching in grasses. Plant Physiol 149:46–55
Ongaro V, Leyser O (2008) Hormonal control of shoot branching. J Exp Bot 59:67–74
Snowden KC, Napoli C (2003) A quantitative study and lateral branching in petunia. Funct Plant Biol 30:987–994
Sorefan K, Booker J, Haurogne K, Goussot M, Bainbridge K, Foo E et al (2003) MAX4 and RMS1 are orthologous dioxygenaselike genes that regulate shoot branching in Arabidopsis and pea. Genes Dev 17:1469–1474
Stirnberg P, Van De Sande K, Leyser HMO (2002) MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Dev 129:1131–1141
Takeda K (1997) Internode elongation and dwarfism in some gramineous plants. Gamma Field Sym 16:l–18
Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M et al (2003) The OsTB1 gene negatively regulates lateral branching in rice. Plant J 33:513–520
Turnbull CGN, Booker JP, Leyser HMO (2002) Micrografting techniques for testing long–distance signalling in Arabidopsis. Plant J 32:255–262
Wang Y, Li J (2008) Molecular basis of plant architecture. Annu Rev Plant Biol 59:253–279
Zhao JF, Wang T, Wang MX, Liu YY, Yuan SJ, Gao YN et al (2013) DWARF3 participates in an SCF complex and associates with DWARF14 to suppress rice shoot branching. Plant Cell Physiol 55:1096–1109
Zhou F, Lin Q, Zhu L, Ren Y, Zhou K, Shabek N et al (2013) D14–SCFD3–dependent degradation of D53 regulates strigolactone signalling. Nature 504:406
Zou J, Chen Z, Shang Z, Zhang W, Jiang G, Zhao X et al (2005) Characterizations and fine mapping of a mutantgene for high tillering and dwarf in rice (Oryza sativa L.). Planta 222:604–612
Zou J, Zhang S, Zhang W, Li G, Chen Z, Zhai W et al (2006) The rice HIGH–TILLERING DWARF1 encoding anortholog of Arabidopsis MAX3 is required for negative regulation of the outgrowth of axillary buds. Plant J 48:687–696
Acknowledgments
This work was supported by the Science and Technology Office of Zhejiang Province (2016C32085 and 2012C12901-2), Open Foundation of Key Laboratory for Crop Germplasm of Zhejiang Province and the Program for Innovative Research Team in University (IRT1185).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest to this work and have no financial and personal relationships with other people or organizations that can inappropriately influence their work, there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the position presented in, or the review of, the manuscript entitled.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liang, R., Qin, R., Yang, C. et al. Identification and Characterization of a Novel Strigolactone-Insensitive Mutant, Dwarfism with High Tillering Ability 34 (dhta-34) in Rice (Oryza sativa L.). Biochem Genet 57, 403–420 (2019). https://doi.org/10.1007/s10528-018-9896-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-018-9896-z