Abstract
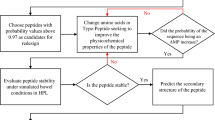
Antimicrobial peptides (AMPs) are promising cationic and amphipathic molecules to fight antibiotic resistance. To search for novel AMPs, we applied a computational strategy to identify peptide sequences within the organisms' proteome, including in-house developed software and artificial intelligence tools. After analyzing 150.450 proteins from eight proteomes of bacteria, plants, a protist, and a nematode, nine peptides were selected and modified to increase their antimicrobial potential. The 18 resulting peptides were validated by bioassays with four pathogenic bacterial species, one yeast species, and two cancer cell-lines. Fourteen of the 18 tested peptides were antimicrobial, with minimum inhibitory concentrations (MICs) values under 10 µM against at least three bacterial species; seven were active against Candida albicans with MICs values under 10 µM; six had a therapeutic index above 20; two peptides were active against A549 cells, and eight were active against MCF-7 cells under 30 µM. This study's most active antimicrobial peptides damage the bacterial cell membrane, including grooves, dents, membrane wrinkling, cell destruction, and leakage of cytoplasmic material. The results confirm that the proposed approach, which uses bioinformatic tools and rational modifications, is highly efficient and allows the discovery, with high accuracy, of potent AMPs encrypted in proteins.


Similar content being viewed by others
References
Bacalum M, Radu M (2015) Cationic antimicrobial peptides cytotoxicity on mammalian cells: an analysis using therapeutic index integrative concept. Int J Pept Res Ther 21:47–55. https://doi.org/10.1007/s10989-014-9430-z
Bhullar K, Waglechner N, Pawlowski A et al (2012) Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 7(4):e34953. https://doi.org/10.1371/JOURNAL.PONE.0034953
Boman HG (2003) Antibacterial peptides: Basic facts and emerging concepts. J Intern Med 254:197–215. https://doi.org/10.1046/j.1365-2796.2003.01228.x
Branch J, Orduz S, Mera C et al. (2022) AmpClass 1.0. Una herramienta informática para la predicción de la actividad antimicrobiana de péptidos. Dirección Nacional de Derechos de Autor. Registro 13-92-95, Colombia
Brand GD, Ramada MHS, Manickchand JR et al (2019) Intragenic antimicrobial peptides (IAPs) from human proteins with potent antimicrobial and anti-inflammatory activity. PLoS ONE 14(8):e0220656. https://doi.org/10.1371/JOURNAL.PONE.0220656
D’Costa VM, King CE, Kalan L et al (2011) Antibiotic resistance is ancient. Nature 477:457–461. https://doi.org/10.1038/nature10388
Deslouches B, Di YP (2017) Oncotarget 46635 www.impactjournals.com/oncotarget antimicrobial peptides with selective antitumor mechanisms: prospect for anticancer applications. Oncotarget 8:46635–46651
Duque-Salazar G, Mendez-Otalvaro E, Ceballos-Arroyo AM, Orduz S (2020) Design of antimicrobial and cytolytic peptides by computational analysis of bacterial, algal, and invertebrate proteomes. Amino Acids 52:1403–1412. https://doi.org/10.1007/S00726-020-02900-W/TABLES/4
Felício MR, Silva ON, Gonçalves S et al (2017) Peptides with dual antimicrobial and anticancer activities. Front Chem 5:1–9. https://doi.org/10.3389/fchem.2017.00005
Fry DE (2018) Antimicrobial peptides. Surg Infect (larchmt) 19:804–811. https://doi.org/10.1089/sur.2018.194
Gautier R, Douguet D, Antonny B, Drin G (2008) HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics 24:2101–2102. https://doi.org/10.1093/BIOINFORMATICS/BTN392
Gómez E, Orduz S (2017) PepMultiFinder 1.0: Un algoritmo para buscar péptidos bioactivos en proteomas o listas de secuencias de proteínas
Hincapié O, Giraldo P, Orduz S (2018) In silico design of polycationic antimicrobial peptides active against Pseudomonas aeruginosa and Staphylococcus aureus. Antonie Van Leeuwenhoek. Int J Gen Mol Microbiol 111:1871–1882. https://doi.org/10.1007/s10482-018-1080-2
Houyvet B, Zanuttini B, Corre E et al (2018) Design of antimicrobial peptides from a cuttlefish database. Amino Acids 50:1573–1582. https://doi.org/10.1007/s00726-018-2633-4
Lamiable A, Thevenet P, Rey J et al (2016) PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucl Acids Res 44:W449–W454. https://doi.org/10.1093/NAR/GKW329
Li F, Dhordain P, Hearn MTW, Martin LL, Bennett LE (2023) Comparative yields of antimicrobial peptides released from human and cow milk proteins under infant digestion conditions predicted by in silico methodology. Food Funct 14(11):5442–5452. https://doi.org/10.1039/d3fo00748k
Liu L, Wang C, Zhang M, Zhang Z, Wu Y, Zhang Y (2022) An efficient evaluation system accelerates α-helical antimicrobial peptide discovery and its application to global human genome mining. Front Microbiol 13:870361. https://doi.org/10.3389/fmicb.2022.870361
Marcellini L, Giammatteo M, Aimola P, Mangoni ML (2010) Fluorescence and electron microscopy methods for exploring antimicrobial peptides mode(s) of action. Methods Mol Biol 618:249–266. https://doi.org/10.1007/978-1-60761-594-1_16/COVER
Mooney C, Haslam NJ, Holton TA et al (2013) PeptideLocator: prediction of bioactive peptides in protein sequences. Bioinformatics 29:1120–1126. https://doi.org/10.1093/bioinformatics/btt103
Mulani MS, Kamble EE, Kumkar SN et al (2019) Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol 10:539. https://doi.org/10.3389/fmicb.2019.00539
Neundorf I (2019) Antimicrobial and cell-penetrating peptides: how to understand two distinct functions despite similar physicochemical properties. Adv Exp Med Biol 1117:93–109. https://doi.org/10.1007/978-981-13-3588-4_7
O’Driscoll NH, Labovitiadi O, Cushnie TPT et al (2013) Production and evaluation of an antimicrobial peptide-containing wafer formulation for topical application. Curr Microbiol 66:271–278. https://doi.org/10.1007/S00284-012-0268-3/METRICS
Perron GG, Whyte L, Turnbaugh PJ et al (2015) Functional characterization of bacteria isolated from ancient Arctic soil exposes diverse resistance mechanisms to modern antibiotics. PLoS ONE 10(3):e0069533. https://doi.org/10.1371/JOURNAL.PONE.0069533
Pinilla G, Coronado YT, Chaves G et al (2022) In vitro antifungal activity of LL-37 analogue peptides against Candida spp. J Fungi 8:1173. https://doi.org/10.3390/JOF8111173
Santos MA, Silva FL, Lira BOV, Cardozo Fh JL, Vasconcelos AG, Araujo AR, Murad AM, Garay AV, Freitas SM, Leite JRSA, Bloch C Jr, Ramada MHS, de Oliveira AL, Brand GD (2023) Probing human proteins for short encrypted antimicrobial peptides reveals Hs10, a peptide with selective activity for gram-negative bacteria. Biochim Biophys Acta Gen Subj 1867:130265. https://doi.org/10.1016/j.bbagen.2022.130265
Strandberg E, Tiltak D, Ieronimo M et al (2007) Influence of C-terminal amidation on the antimicrobial and hemolytic activities of cationic α-helical peptides. Pure Appl Chem 79:717–728. https://doi.org/10.1351/pac200779040717
Tacconelli E, Magrini N (2017) Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organ Essent Med Heal Prod 1–7
Tamayo S, Castañeda C, Orduz S (2018) Type-Peptide 1.0. Dirección Nacional de Derechos de Autor. Registro 13-70-274, Colombia
Torrent M, Andreu D, Nogués VM, Boix E (2011) Connecting peptide physicochemical and antimicrobial properties by a rational prediction model. PLoS ONE 6(2):e16968. https://doi.org/10.1371/JOURNAL.PONE.0016968
Torrent M, Di Tommaso P, Pulido D et al (2012) AMPA: an automated web server for prediction of protein antimicrobial regions. Bioinformatics 28:130–131. https://doi.org/10.1093/bioinformatics/btr604
Torres MDT, Melo MCR, Flowers L, Crescenzi O, Notomista E, de la Fuente-Nunez C (2022) Mining for encrypted peptide antibiotics in the human proteome. Nat Biomed Eng 6(1):67–75. https://doi.org/10.1038/s41551-021-00801-1
Veltri D, Kamath U, Shehu A (2018) Deep learning improves antimicrobial peptide recognition. Bioinformatics 34:2740–2747. https://doi.org/10.1093/bioinformatics/bty179
Waghu FH, Barai RS, Gurung P, Idicula-Thomas S (2016) CAMPR3: a database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res 44:D1094–D1097. https://doi.org/10.1093/nar/gkv1051
Wang G (2017) Antimicrobial peptides: discovery, design, and novel therapeutic strategies. 1–230. https://doi.org/10.1079/9781845936570.0000
Wang G, Li X, Wang Z (2016) APD3: the antimicrobial peptide database as a tool for research and education. Nucl Acids Res 44:D1087–D1093. https://doi.org/10.1093/nar/gkv1278
Wiegand I, Hilpert K (2008) Hancock REW (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 32(3):163–175. https://doi.org/10.1038/nprot.2007.521
World Health Organization (2021) Antimicrobial resistance. In: https://www.who.int/en/news-room/fact-sheets/detail/antimicrobial-resistance
Yang J, Zhang Y (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181. https://doi.org/10.1093/NAR/GKV342
Zhang B, Shi W, Li J et al (2017) Synthesis and biological evaluation of novel peptides based on antimicrobial peptides as potential agents with antitumor and multidrug resistance-reversing activities. Chem Biol Drug Des 90:972–980. https://doi.org/10.1111/CBDD.13023
Funding
This study was supported by Universidad Nacional de Colombia—Sede Medellin, the HERMES research grants numbers 42990 and 55740, and by the Instituto Tecnológico Metropolitano de Medellín (ITM) research grant number PE20202.
Author information
Authors and Affiliations
Contributions
SO, CM, and JB designed the study, worked on data analysis and interpretation, participated in the writing of the manuscript, and reviewed its final version. DM AM and LM participated in the sampling, data collection, laboratory essays, and analysis. They also collaborate on writing the draft of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors declare they have no conflict of interest with the information contained in this manuscript. Also, the authors have no relevant financial or non-financial interests to disclose.
Ethical approval
All procedures followed were in accordance with the ethical standards of the responsible committee.
Informed consent
Informed consent was obtained from the blood donor for being included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Monsalve, D., Mesa, A., Mira, L.M. et al. Antimicrobial peptides designed by computational analysis of proteomes. Antonie van Leeuwenhoek 117, 55 (2024). https://doi.org/10.1007/s10482-024-01946-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10482-024-01946-0