Abstract
An actinobacterial strain A23T, isolated from adult ant Camponotus vagus collected in Ryazan region (Russia) and established as tetracenomycin X producer, was subjected to a polyphasic taxonomic study. Morphological characteristics of this strain included well-branched substrate mycelium and aerial hyphae fragmented into rod-shaped elements. Phylogenetic analyses based on 16S rRNA gene and genome sequences showed that strain A23T was most closely related to Amycolatopsis pretoriensis DSM 44654T. Average nucleotide identity and digital DNA–DNA hybridization values between the genome sequences of isolate A23T and its closest relative, Amycolatopsis pretoriensis DSM 44654T, were 39.5% and 88.6%, which were below the 70% and 95–96% cut-off point recommended for bacterial species demarcation, respectively. The genome size of the isolate A23T was 10,560,374 bp with a DNA G + C content of 71.2%. The whole-cell hydrolysate contained meso-diaminopimelic acid and arabinose and galactose as main diagnostic sugars as well as ribose and rhamnose. It contained MK-9(H4) as the predominant menaquinone and iso-C16:0, iso-C15:0, anteiso-C17:0 and C16:0 as the major cellular fatty acids. Diphosphatidylglycerol and phosphatidylethanolamine prevailed among phospholipids. Mycolic acids were not detected. Based on the phenotypic, genomic and phylogenetic data, isolate A23T represents a novel species of the genus Amycolatopsis, for which the name Amycolatopsis camponoti sp. nov. is proposed, and the type strain is A23T (= DSM 111725T = VKM 2882T).
Similar content being viewed by others
Introduction
The genus Amycolatopsis Lechevalier et al. 1986 belonged to the family Pseudonocardiaceae Embley et al. 1989 (order Pseudonocardiales Labeda and Goodfellow 2015, class Actinomycetia Salam et al. 2020 (Salam et al. 2020), phylum Actinomycetota corrig. Goodfellow 2021 (Oren and Garrity 2021)), and encompasses 85 validly published species names with Amycolatopsis orientalis as its type species (https://lpsn.dsmz.de/genus/amycolatopsis). Amycolatopsis is aerobic to facultatively anaerobic and characterized by branched vegetative hyphae that undergo fragmentation into rod-like and squarish elements. Whole-cell hydrolysates are rich in meso-2,6-diaminopimelic acid, along with arabinose and galactose as whole-cell sugars. The peptidoglycan is of the A1g type. Muramic acid moieties are N-acetylated. Does not contain mycolic acids. The diagnostic phospholipid is phosphatidylethanolamine and/or phosphatidylmethylethanolamine while the occurrence of diphosphatidylglycerol, phosphatidylglycerol, phosphatidylinositol and phosphatidylinositol mannosides is variable (Tan and Goodfellow 2015). Amycolatopsis have been described from diverse environments such as soil, vegetation, human and animal clinical sources, fresh water, rock and subterranean sites (Lee 2009).
The 16S rRNA gene phylogeny provides a useful framework for inferring the relationships between genera in the Pseudonocardiaceae family, but did not seem to have enough taxonomic resolution to distinguish species. The use of multi-locus sequence analysis (MLSA) is able to clarify the taxonomic positions of Amycolatopsis species (Glaeser and Kämpfer 2015). The most reliable way to identify actinobacteria strains is the comparison of genomes from the point of view of DNA–DNA relatedness and construction whole-genome phylogenetic trees (Nouioui et al. 2018).
The studies of 66 publicly available representative genome sequences of Amycolatopsis type strains (https://www.ncbi.nlm.nih.gov/genome/browse#!/prokaryotes/Amycolatopsis) have revealed that Amycolatopsis have comparatively large genomes from nearly 5.62 Mb (Amycolatopsis granulosa DSM 45669T) to 10.94 Mb (Amycolatopsis anabasis EGI 650086T), GC content from 67.8% (Amycolatopsis palatopharingis DSM 44832T) to 72.7% (Amycolatopsis arida DSM 45648T), while median genome size and DNA G + C content are 9.08 Mb and 70.1%, respectively. The circular chromosomes containing over 20 secondary metabolic gene clusters (Kumari et al. 2016).
The genus Amycolatopsis comprises a large group of commercially and medically important actinobacteria capable of producing two major types of antibiotics: glycopeptides and polyketides (Kisil et al. 2021). In this paper, we describe strain A23T, isolated from body of ant Camponotus vagus collected in Ryazan region, Russia. This strain is able to produce the aromatic polyketide antibiotic tetracenomycin X (TcmX) and its new congener 6-hydroxytetracenomycin X (6-OH-Tcm X) which possess antimicrobial and cytotoxic activity (Osterman et al. 2020; Alferova et al. 2021). As recently demonstrated TcmX and 6-OH-Tcm X are potent inhibitors of protein synthesis due to the fact that they bind to the large subunits of prokaryotic or eukaryotic ribosomes, within the polypeptide exit tunnel (Osterman et al. 2020).
Polyphasic study was used to clarify the taxonomic position of novel strain A23T. We propose to establish this strain as a representative for the novel species of the genus Amycolatopsis, with the name Amycolatopsis camponoti sp. nov.
Materials and methods
Collection and microbial isolation
Strain A23T was isolated from bodies of adult workers of Camponotus vagus collected in Kasimovsky District, Ryazan region, Russia (55.01138 N, 41.73078 E) (Zakalyukina et al. 2021). Five individuals were washed three times in sterile distilled water and then crushed by tissue microhomogenizer with sterile saline solution. Aliquots of this mixtures were spread over M490 medium (HiMediaLab) supplemented with nystatin and nalidixic acid at final concentrations of 250 μg/mL and 10 μg/mL, respectively, and incubated for 14 days at 28 °C (Zakalyukina et al. 2019). The strain was purified and maintained on Organic medium 79 (Prauser and Falta 1968), and preserved as suspension of mycelial fragments and spores in 20% (v/v) glycerol at − 20 °C.
Genome features and phylogenomic analysis
Genome of strain A23T was sequenced de novo by Novogene Co., Ltd. (https://en.novogene.com/), using the Illumina NovaSeq 6000 platform and fully annotated using RAST prokaryotic genome annotation service (https://rast.nmpdr.org/) and submitted in GenBank (assembly accession: GCA_902497555.1) (Osterman et al. 2020).
Average nucleotide identity (ANI) (Rodríguez-R and Konstantinidis 2016) and in silico digital DNA:DNA hybridization (DDH) were calculated using JSpecies WS (http://jspecies.ribohost.com/jspeciesws/), and GGDC method, with the recommended formula 2, available at the TYGS web service, respectively (Meier-Kolthoff and Göker 2019).
Phylogenomic analysis was performed using Type (Strain) Genome Server (https://tygs.dsmz.de/). The phylogenomic tree inferred with FastME 2.1.6.1 (Lefort et al. 2015) from GBDP distances calculated from genome sequences. The branch lengths are scaled in terms of GBDP distance formula d5.
16S rRNA phylogeny
The full-length 16S rRNA gene sequences of strain A23T was extracted from the whole genome sequence (CABVGP010000001.1) and was compared to sequences of type strains in the EzBioCloud database (www.ezbiocloud.net).
Evolutionary trees based on 16S rRNA gene sequences were inferred with the neighbour-joining, maximum-parsimony and maximum-likelihood tree-making algorithms after CLUSTAL W alignment by using MEGA software version X (Kumar et al. 2018) (https://www.megasoftware.net). These evolutionary analyzes involved 24 nucleotide sequences. All positions with less than 95% site coverage were eliminated, i.e., fewer than 5% alignment gaps, missing data, and ambiguous bases were allowed at any position (partial deletion option). There were a total of 1414 positions in the final dataset.
Phenotypic characterization
Cultural characteristics of strain A23T were observed on the range of ISP 2-ISP 7 media (Shirling and Gottlieb 1966), Organic medium 79 and modified Bennett’s agar (Tan et al. 2006b) after cultivation up to 14 days at 28 °C. Cell morphology of strain A23T on Organic medium 79 after cultivation at 28 °C for 10 days was studied using scanning electron microscopy (JSM-6380LA, JEOL). Motility test was carried out using the “hanging drop” method by light microscope (Fisherbrand AX-502, Thermo Fisher Scientific).
Carbon source utilization was assessed on basal medium ISP 9 (Shirling and Gottlieb 1966) with addition of 0.04% solution of bromocresol purple at 28 °C for 14 days. Enzyme activities were estimated using paper indicator system (NPO Microgen, Russia) according to the manufacturer’s recommendations at 28 °C for 7 days. The degradation of casein, starch and cellulose was estimated on clearing of the insoluble compounds around areas of growth (Williams et al. 1983). The ability of strain to grow at a different pH (pH 5.0, pH 6.0, pH 7.0, pH 8.0, pH 9.0) was tested on a modified ISP 9 medium (g/L: (NH4)2SO4–2.64, MgSO4 × 7H2O–1, glucose—10, agar—20), buffered with phosphate solutions according to Williams et al. (Williams et al. 1971), at 28 °C for 14 days. The growth at different range of temperature (5 °C, 10 °C, 20 °C, 30 °C, 40 °C) and salinity (1%, 5%, 8%) were assessed on Organic medium 79 after 14 days of incubation.
Chemotaxonomy
Cell biomass of strains A23T and A. pretoriensis DSM 44654T were obtained from cultures grown in DSMZ 554 broth medium and on a rotary shaker (180 r.p.m) at 28 °C. After 120-h growth cells were centrifuged, washed three times in sterile distilled water and freeze-dried. Whole-cell sugars (Lechevalier and Lechevalier 1970; Staneck and Roberts 1974), menaquinone (Collins et al. 1985) and diaminopimelic acids analyses (Schleifer and Kandler 1972) were carried out. Standard chromatographic procedures were used to determine polar lipid pattern for strain A23T and A. pretoriensis DSM 44654T following the protocol of Minnikin et al. (1984).
Cellular fatty acids extracts were prepared using minor modifications of the protocol of Miller (Miller 1982) and Kuykendall et al. (Kuykendall et al. 1988). Gas chromatography (Agilent 6890N instrument) was used to analyse the fatty acid methyl esters which were identified using Sherlock Microbial Identification system (MIDI, Microbial ID, Newark, DE 19711 U.S.A.) and the Actin6 database (Sasser 1990). The analysis was supplemented by a GC-MS run on an Agilent GC-MS 7000D for identity confirmation. Mycolic acids extraction was performed according to Vilchéze and Jacobs (Vilcheze and Jacobs 2007). Cell lysis was carried out in KOH/MeOH solution at 95 °C overnight and extracted with chloroform. Dried extracts were recovered in chloroform:MeOH (9:1) and analyzed in negative ion mode on an Agilent QTof mass spectrometer by direct infusion into the ESI source (300 µL/min). Mycolic acids were identified by comparison of the exact masses of known mycolic acid structures to the measured ones (Bouam et al. 2018).
Analysis of bioactive compound biosynthetic gene clusters and potential pathogenicity
Secondary metabolite biosynthetic gene clusters in complete genome strain A23T (CABVGP010000000) and A. pretoriensis DSM 44654T (GenBank Accessions FNUJ01000000) were identified with the bacterial version of antiSMASH 6.1.0 (https://antismash.secondarymetabolites.org/). Homologous regions on each genome were identified using NCBI Blastn (https://blast.ncbi.nlm.nih).
For predicting the pathogenicity of A23T and closely related strains we used PathogenFinder (http://cge.cbs.dtu.dk/services/PathogenFinder/), a web-server for the prediction of bacterial pathogenicity by analysing the input genome (Cosentino et al. 2013).
Results and discussion
Amycolatopsis strains are well known as producers for the commercially used antibiotics, e.g. vancomycin, rifamycin, eremomycin (Kisil et al. 2021). Other compounds with antibacterial, antifungal or antiviral properties that have been derived from Amycolatopsis strains are quartromycin, octacosamicin, chelocardin, kigamicin and the macrotermycins A–D (Chen et al. 2016; Kumari et al. 2016; Beemelmanns et al. 2017). It has been shown that there is a relationship between the distribution of biosynthetic gene clusters (BGCs) and phylogenetic lineages in Amycolatopsis genomes. Amycolatopsis strains that produce, or have the potential to produce, a particular class of antibiotic are phylogenetically related (Adamek et al. 2018). Strain A23T was previously known to produce bioactive compounds (Osterman et al. 2020; Alferova et al. 2021) was subjected to a polyphasic taxonomic approach to clarify its taxonomic position.
Phylogenomic analysis based on whole-genome sequences showed that strain A23T formed a well-supported monophyletic clade with A. pretoriensis DSM 44654T with 100% bootstrap value (Fig. 1). The neighbor-joining (Fig. S1), maximum-likelihood (Fig. S2) and maximum-parsimony (Fig. S3) trees based on 16S rRNA full-length gene sequences revealed that strain A23T formed a subclade in the Amycolatopsis tree together with Amycolatopsis pretoriensis DSM 44654T and Amycolatopsis lexingtonensis NRRL B-24131T, concurrently the similarity detected by EzBioCloud is 99.1% and 99.2% respectively. The isolate was also found to share relatively high 16S rRNA gene similarities with the type strains of A. rifamycinica DSM 46095T (99.3%), A. kentuckyensis NRRL B-24129T (99.1%), A. tolypomycina DSM 44544T (98.9%), A. vancoresmycina DSM 44592T (98.8%), A. eburnea NBRC 113658T (98.6%), A. balhimycina DSM 44591T (98.5%), A. mediterranei NRRL B-3240T (98.4%), A. vastitatis NRRL B-65279T (98.3%) and A. australiensis DSM 44671T (97.9%).
Previous phylogenomic analyses revealed that the Amycolatopsis clade is divided into four major phylogenomic subclades (Adamek et al. 2018; Sangal et al. 2018). This study showed that strain A23T is closely related to type strains within the group B subclades sensu Adamek et al. 2018 and Sangal et al. 2018. The study of Sánchez-Hidalgo with colleagues (Sánchez-Hidalgo et al. 2018) showed 3 major clades and 11 groups. The strains closely related to strain A23T are within subclade AOS, group C sensu Sánchez-Hidalgo et al. 2018. The genome-based phylogenies of Adamek et al. 2018 and Sánchez-Hidalgo et al. 2018 were based on MLSA, while Sangal et al. 2018 and Teo et al 2021 were based on core protein sequences/core-proteome. The phylogenomic analysis presented in this study, based on GBDP distances calculated from genome sequences, showed consistency with previous studies (Fig. 1).
The complete genome size of strain A23T was 10,560,374 bp with DNA G + C content of 71.2%, which was consistent with the G + C content of the genus Amycolatopsis (Teo et al. 2021). Similar genome features were observed for the closest neighbor A. pretoriensis DSM 44654T (genome size 10,299,026 bp and G + C content 71.2%). Representatives of the group B are characterized by the presence of a high number of biosynthetic gene clusters ranged from 28 to 41 per genome (Adamek et al. 2018). That fact explains the ability of majority of this clade to produce antibiotics and/or bioactive molecules (Tan and Goodfellow 2015). The genome properties of strain A23T and other type strains within group B sensu Adamek et al. 2018 and Sangal et al. 2018 are summarized in Table 1.
The ANI and in silico DDH values between strain A23T and strain DSM 44654T were 39.5% and 88.6%, respectively. The ANI and in silico DDH values between strain A23T and other related species of the genus Amycolatopsis were below the recommended thresholds of 95–96% and 70% for species demarcation (Ciufo et al. 2018; Meier-Kolthoff and Göker 2019) (Table S1).
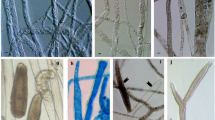
Cells of strain A23T were aerobic, Gram-positive, non-motile. This strain showed a good growth on ISP 2-ISP 5 and moderate on both ISP 6 and ISP 7. The colours of substrate mycelia varied from light ivory to sulfur yellow while aerial hyphae usually were white or cream (Table 2). The branching mycelia (diameter 0.42 µm) fragmented into different lengths rod-shaped elements (Fig. 2), which is typically observed for Amycolatopsis (Franco and Labeda 2014).
Strain A23T, as well as its closely related A. australiensis DSM 44671T, A. balhimycina DSM 44591T, A. eburnea NBRC 113658T, A. kentuckyensis NRRL B-24129T, A. lexingtonensis NRRL B-24131T, A. mediterranei NRRL B-3240T, A. pretoriensis DSM 44654T, A. rifamycinica DSM 46095T, A. tolypomycina DSM 44544T, A. vancoresmycina DSM 44592T, A. vastitatis NRRL B-65279T, produced acid from glucose and inositol. All the above mentioned strains were capable of peptonization of gelatin (Table 3). The majority of considered strains including strain A23T were able to utilize arabinose, fructose, galactose, sucrose and decompose of urea. They grew at 30 °C and in the presence of NaCl 1% w/v, but none produced amylases. The optimum growth temperature and pH of strain A23T were 28–30 °C and pH 7, accordingly, but it was able to grow between pH 6.0–9.0 and up to NaCl 5.0% (w/v).
The whole-cell hydrolysates of strain A23T contained meso-2,6-diaminopimelic acid, arabinose, galactose, ribose and a trace of rhamnose as whole-cell sugars. However, strain DSM 44654T had arabinose and galactose as the major cell sugars while ribose and rhamnose were present in traces. No mycolic acid was detected. Both strains showed similar isoprenoid quinone profile with MK-9(H4) as the predominant one (Table S3). The major fatty acids (> 10%) for strain A23T were iso-C16:0, iso-C15:0, anteiso-C17:0 and C16:0 while the type strain of A. pretoriensis species had iso-C16:0 and iso-C15:0 (Table S4). The polar lipid pattern of A23T included diphosphatidylglycerol, phosphatidylethanolamine (diagnostic lipid), aminophospholipid, a glycolipid, phospholipid and unidentified lipids (Fig. S6), as well as for strain DSM 44654T (Fig. S7).
Online tool antiSMASH predicted 32 secondary metabolite gene clusters in the genome of strain A23T (Table S5) and 30 ones in A. pretoriensis (Table S6). In total, from A23T genome it was identified 11 PKS clusters, 10 NRPS and gene NRPS-like clusters, 2 RiPPs and 4 terpenes, while from DSM 44654T there were 10, 10, 2 and 4 accordingly.
The genome mining of strain A23T revealed that this strain has the potential to produce wide range of secondary metabolites including limazepines A, C-F and macrotermycins A-D (Table S5). The limazepines belong to the growing group of the pyrrolo[1,4]benzodiazepine antitumor antibiotics. Their inherent antitumor and antibacterial activities are due to their ability to regulate gene expression by recognizing and binding with DNA GC base pairs (Fotso et al. 2009). These limazepines’ clusters were identical to that in strain Streptomyces sp. ICBB 8177, and showed also 82% of genes similarity with the sequence of tomaymycin in strain Streptomyces achromogenes (Li et al. 2009) (Fig. 3). Macrotermycin biosynthetic gene clusters with 96% similarity to that of Amycolatopsis strain sp. M39 (Beemelmanns et al. 2017) were found in the genome of strains A23T (Fig. S8). Macrotermycins A-D are 20-membered glycosylated polyketide macrolactams’s had antibacterial and also selective antifungal activity and were isolated from a termite-associated actinomycete, Amycolatopsis sp. M39 (Beemelmanns et al. 2017).
The DSM 44654T genome contained the same clusters encoded limazepines and macrotermycins, supporting the view that phylogenetic similarity implies the presence of closely related biosynthetic pathways (Adamek et al. 2018).
Previously described tetracenomycin biosynthesis gene cluster and its paralogus Tcm2 cluster (Osterman et al. 2020) were detected in 1.7 and 2.5 regions of genome A23T accordingly. Among all related strains A. rifamycinica DSM 46095T and A. balhimycina DSM 44591T have orthologous clusters in their genomes (Osterman et al. 2020). Despite the relatively closeness, no Tcm clusters were detected in the genome sequence of strain DSM 44654T and other related strains.
The vast majority of Amycolatopsis from B-group were predicted as non human pathogen, the values of probability of being a human pathogen were below one (Table S7). Nevertheless, the protein families associated with pathogenicity were detected, so ABC-type transport system protein of Saccharomonospora viridis DSM 43017 was present in the genome of A23T and all the strains examined (Table S7).
Based on phenotypic, phylogenetic and genomics analyses, strain A23T, producer of a valuable substance tetracenomycin X, is considered as a type strain of a novel species with the proposed name, Amycolatopsis camponoti.
Description of Amycolatopsis camponoti sp. nov.
Amycolatopsis camponoti (cam.po.no′ti. N.L. gen. n. camponoti of Camponotus, referring to the insect Camponotus vagus Scopoli, from which the type strain was isolated).
Aerobic, Gram-strain-positive, non-motile and filamentous actinobacteria. The aerial mycelia fragments into rod-shaped fragments (0.42 µm in diameter). Well-developed substrate mycelium varies from light ivory to sulfur yellow, and the colour of aerial mycelium usually is white on ISP 2-ISP 4, ISP 6, MBA and Organic 79 media. When growing for three weeks in liquid Organic medium 79, it produces soluble pigments that ranges from faintly brown to red.
The optimum growth temperature and pH are 28–30 °C and pH 7, but it is unable to grow at 10 and 40 °C and out of range 6–9 pH same as above 5.0% salinity (w/v). It metabolizes arabinose, fructose, galactose, inositol, lactose, maltose, mannitol, rhamnose, sorbitol, sucrose, xylose and weakly raffinose, but unable to use adonitol, cellulose, starch and salicin. Strain A23T demonstrates noticeable activity of β-glucosidase, arginine dihydrolase, lysine and ornithine decarboxylases.
The cell wall contains meso-2,6-diaminopimelic acid, arabinose, galactose, ribose and a trace of rhamnose as cell sugars. Major cellular fatty acids are iso-C16:0, iso-C15:0, anteiso-C17:0 and C16:0. The predominant menaquinone is MK-9(H4), while MK-9(H2) and MK-8(H4) are present as minor components.
The type strain is A23T (= DSM 111725T = VKM Ac-2882T), isolated from bodies of ants Camponotus vagus in Ryazan region, Russia. The genome size of the isolate A23T is 10,560,374 bp with a DNA G + C content of 71.2%. The GenBank accession number for the 16S rRNA gene sequence and the genome assembly of strain A23T are KY952635.2 and GCA_902497555, respectively.
Data availability statement
The GenBank accession number for the 16S rRNA gene sequence and the genome assembly of strain A23T are KY952635.2 and GCA_902497555, respectively. Detailed data on genome analysis A23T are presented IMG/M DataBase (https://gold.jgi.doe.gov/).
Change history
22 March 2023
A Correction to this paper has been published: https://doi.org/10.1007/s10482-023-01815-2
10 April 2022
A Correction to this paper has been published: https://doi.org/10.1007/s10482-022-01729-5
References
Adamek M, Alanjary M, Sales-Ortells H et al (2018) Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis species. BMC Genom 19:426. https://doi.org/10.1186/s12864-018-4809-4
Alferova VA, Maviza TP, Biryukov MV et al (2021) Biological evaluation and spectral characterization of a novel tetracenomycin X congener. Biochimie. https://doi.org/10.1016/J.BIOCHI.2021.09.014
Bala S, Khanna R, Dadhwal M et al (2004) Reclassification of Amycolatopsis mediterranei DSM 46095 as Amycolatopsis rifamycinica sp. nov. Int J Syst Evol Microbiol 54:1145–1149. https://doi.org/10.1099/ijs.0.02901-0
Beemelmanns C, Ramadhar TR, Kim KH et al (2017) Macrotermycins A-D, glycosylated macrolactams from a termite-Associated Amycolatopsis sp. M39. Org Lett. https://doi.org/10.1021/acs.orglett.6b03831
Bouam A, Armstrong N, Levasseur A, Drancourt M (2018) Mycobacterium terramassiliense, Mycobacterium rhizamassiliense and Mycobacterium numidiamassiliense sp. nov., three new Mycobacterium simiae complex species cultured from plant roots. Sci Rep 8:9309. https://doi.org/10.1038/s41598-018-27629-1
Chaiya L, Matsumoto A, Wink J et al (2019) Amycolatopsis eburnea sp. Nov., an actinomycete associated with arbuscular mycorrhizal fungal spores. Int J Syst Evol Microbiol 69:3603–3608. https://doi.org/10.1099/ijsem.0.003669
Chen S, Wu Q, Shen Q, Wang H (2016) Progress in understanding the genetic information and biosynthetic pathways behind Amycolatopsis antibiotics, with implications for the continued discovery of novel drugs. ChemBioChem 17:119–128. https://doi.org/10.1002/cbic.201500542
Ciufo S, Kannan S, Sharma S et al (2018) Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int J Syst Evol Microbiol 68:2386–2392. https://doi.org/10.1099/ijsem.0.002809
Collins MD, Goodfellow M, Minnikin DE, Alderson G (1985) Menaquinone composition of mycolic acid-containing actinomycetes and some sporoactinomycetes. J Appl Bacteriol. https://doi.org/10.1111/j.1365-2672.1985.tb01431.x
Cosentino S, Larsen MV, Aarestrup FM, Lund O (2013) PathogenFinder—distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE 8:e77302. https://doi.org/10.1371/journal.pone.0077302
Fotso S, Zabriskie TM, Proteau PJ et al (2009) Limazepines A−F, Pyrrolo[1,4]benzodiazepine antibiotics from an Indonesian Micrococcus sp. J Nat Prod. https://doi.org/10.1021/np800827w
Franco CMM, Labeda DP (2014) The Order Pseudonocardiales. In: The prokaryotes. Springer, Berlin
Glaeser SP, Kämpfer P (2015) Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst Appl Microbiol 38:237–245. https://doi.org/10.1016/J.SYAPM.2015.03.007
Idris H, Nouioui I, Pathom-aree W et al (2018) Amycolatopsis vastitatis sp. nov., an isolate from a high altitude subsurface soil on Cerro Chajnantor, northern Chile. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-018-1039-3
Kisil OV, Efimenko TA, Efremenkova OV (2021) Looking back to Amycolatopsis: history of the antibiotic discovery and future prospects. Antibiotics. https://doi.org/10.3390/antibiotics10101254
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Kumari R, Singh P, Lal R (2016) Genetics and genomics of the genus Amycolatopsis. Indian J Microbiol 56:233–246. https://doi.org/10.1007/s12088-016-0590-8
Kuykendall LD, Roy MA, O’Neill JJ, Devine TE (1988) Fatty acids, antibiotic resistance, and deoxyribonucleic acid homology groups of Bradyrhizobium japonicum. Int J Syst Bacteriol 38:358–361. https://doi.org/10.1099/00207713-38-4-358
Labeda DP, Donahue JM, Williams NM et al (2003) Amycolatopsis kentuckyensis sp. nov., Amycolatopsis lexingtonensis sp. nov. and Amycolatopsis pretoriensis sp. nov., isolated from equine placentas. Int J Syst Evol Microbiol 53:1601–1605. https://doi.org/10.1099/ijs.0.02691-0
Lechevalier MP, Lechevalier HA (1970) Chemical composition as a criterion in the classification of aerobic actinomycetes. Int J Syst Evol Microbiol 20:435–443
Lechevalier MP, Prauser H, Labeda DP, Ruan JS (1986) Two new genera of nocardioform actinomycetes: Amycolata gen. nov. and Amycolatopsis gen. nov. Int J Syst Bacteriol 36:29–37. https://doi.org/10.1099/00207713-36-1-29
Lee SD (2009) Amycolatopsis ultiminotia sp. nov., isolated from rhizosphere soil, and emended description of the genus Amycolatopsis. Int J Syst Evol Microbiol 59:1401–1404. https://doi.org/10.1099/ijs.0.006577-0
Lefort V, Desper R, Gascuel O (2015) FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol Biol Evol 32:2798–2800. https://doi.org/10.1093/molbev/msv150
Li W, Chou S, Khullar A, Gerratana B (2009) Cloning and characterization of the biosynthetic gene cluster for tomaymycin, an SJG-136 monomeric analog. Appl Environ Microbiol. https://doi.org/10.1128/AEM.02325-08
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182. https://doi.org/10.1038/s41467-019-10210-3
Miller LT (1982) Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J Clin Microbiol 16:584–586
Minnikin DE, O’Donnell AG, Goodfellow M, et al (1984) An Integrated Procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Nouioui I, Carro L, García-López M et al (2018) Genome-based taxonomic classification of the phylum Actinobacteria. Front Microbiol 9:2007. https://doi.org/10.3389/fmicb.2018.02007
Oren A, Garrity GMY (2021) Valid publication of the names of forty-two phyla of prokaryotes. Int J Syst Evol Microbiol 71:005056. https://doi.org/10.1099/ijsem.0.005056
Osterman IA, Wieland M, Maviza TP et al (2020) Tetracenomycin X inhibits translation by binding within the ribosomal exit tunnel. Nat Chem Biol. https://doi.org/10.1038/s41589-020-0578-x
Prauser H, Falta R (1968) Phagensensibilität, Zellwand-Zusammensetzung und Taxonomie von Actinomyceten. J Basic Microbiol 8:39–46
Rodríguez-RL, Konstantinidis K (2016) The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes
Salam N, Jiao J-Y, Zhang X-T, Li W-J (2020) Update on the classification of higher ranks in the phylum Actinobacteria. Int J Syst Evol Microbiol 70:1331–1355. https://doi.org/10.1099/ijsem.0.003920
Sánchez-Hidalgo M, González I, Díaz-Muñoz C et al (2018) Comparative genomics and biosynthetic potential analysis of two lichen-isolated Amycolatopsis strains. Front Microbiol 9:1–16. https://doi.org/10.3389/fmicb.2018.00369
Sangal V, Goodfellow M, Blom J et al (2018) Revisiting the taxonomic status of the biomedically and industrially important genus Amycolatopsis, using a phylogenomic approach. Front Microbiol 9:2281. https://doi.org/10.3389/fmicb.2018.02281
Sasser M (1990) Bacterial identification by gas chromatographic analysis of fatty acids methyl esters (GC-FAME). MIDI Labs Inc, Newark
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. https://doi.org/10.1128/br.36.4.407-477.1972
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340. https://doi.org/10.1099/00207713-16-3-313
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol. https://doi.org/10.1128/am.28.2.226-231.1974
Tan GYA, Goodfellow M (2015) Amycolatopsis. In: Bergey’s manual of systematics of archaea and bacteria. John Wiley & Sons, Ltd, pp 1–40
Tan GYA, Robinson S, Lacey E, Goodfellow M (2006a) Amycolatopsis australiensis sp. nov., an actinomycete isolated from arid soils. Int J Syst Evol Microbiol 56:2297–2301. https://doi.org/10.1099/ijs.0.64260-0
Tan GYA, Ward AC, Goodfellow M (2006b) Exploration of Amycolatopsis diversity in soil using genus-specific primers and novel selective media. Syst Appl Microbiol 29:557–569. https://doi.org/10.1016/j.syapm.2006.01.007
Teo WFA, Tan GYA, Li W-J (2021) Taxonomic note on the family Pseudonocardiaceae based on phylogenomic analysis and descriptions of Allosaccharopolyspora gen. nov. and Halosaccharopolyspora gen. nov. Int J Syst Evolut Microbiol 71:005075. https://doi.org/10.1099/ijsem.0.005075
Vilcheze C, Jacobs WR (2007) Isolation and analysis of Mycobacterium tuberculosis mycolic acids. Curr Protoc Microbiol 5:10A–3
Williams S, Davies FL, Mayfield C, Khan MR (1971) Studies on the ecology of actinomycetes in soil II. The pH requirements of streptomycetes from two acid soils. Soil Biol Biochem. https://doi.org/10.1016/0038-0717(71)90014-9
Williams ST, Goodfellow M, Alderson G et al (1983) Numerical classification of streptomyces and related genera. Microbiology 129:1743–1813. https://doi.org/10.1099/00221287-129-6-1743
Wink JM, Kroppenstedt RM, Ganguli BN et al (2003) Three new antibiotic producing species of the genus Amycolatopsis, Amycolatopsis balhimycina sp. nov., A. tolypomycina sp. nov., A. vancoresmycina sp. nov., and description of Amycolatopsis keratiniphila subsp. keratiniphila subsp. nov. and A. keratiniphi. Syst Appl Microbiol 26:38–46
Zakalyukina YV, Birykov MV, Lukianov DA et al (2019) Nybomycin-producing Streptomyces isolated from carpenter ant Camponotus vagus. Biochimie. https://doi.org/10.1016/j.biochi.2019.02.010
Zakalyukina YV, Zaytsev AR, Biryukov MV (2021) Study of cellulose-destroying activity of actinobacteria associated with ants. Mosc Univ Biol Sci Bull. https://doi.org/10.3103/S0096392521010065
Acknowledgements
The authors are indebted to Marlen Jando and Gabriele Pötter (Leibniz Institute DSMZ–German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany) for support with the growth cultures. SEM studies were carried out at the Shared Research Facility “Electron microscopy in life sciences” at Moscow State University (Unique Equipment “Three-dimensional electron microscopy and spectroscopy”). The authors acknowledge partial support from M.V.Lomonosov Moscow State University Program of Development. The authors are grateful to anonymous reviewers for improving the clarity and quality of the manuscript.
Funding
This research was funded by Sirius University, project [BTH-RND-2127] for YVZ and by Ministry of Science and Higher Education of the Russian Federation (Agreement No 075–15-2021-1085) for IAO, MVB.
Author information
Authors and Affiliations
Contributions
YVZ carried out the experiments, visualization, analysis the data, and writing draft manuscript, IAO conceptualized the project, JW, MN-S carried out chemotaxonomical assays, IN corrected and reviewed the draft, MVB supervised the project. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
This article does not contain any studies with human participants and/or animals performed by any of the authors. The formal consent is not required in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: The number of the strain in VKM was incorrectly published as “VKM 2882”, the correct number is “VKM Ac-2882”. The missing funding information “ This research was funded by Sirius University, project [BTH-RND-2127] for YVZ and by Ministry of Science and Higher Education of the Russian Federation (Agreement № 075–15-2021-1085) for IAO, MVB” are included.
The VKM culture collection number was corrected in the published article. However, to comply with the rules of the International Code of Nomenclature of Prokaryotes (ICNP), the author would like to reverse the correction updated in the original article and issue a corrigendum containing the full corrected protologue. The original version of the article is restored.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zakalyukina, Y.V., Osterman, I.A., Wolf, J. et al. Amycolatopsis camponoti sp. nov., new tetracenomycin-producing actinomycete isolated from carpenter ant Camponotus vagus. Antonie van Leeuwenhoek 115, 533–544 (2022). https://doi.org/10.1007/s10482-022-01716-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-022-01716-w