Abstract
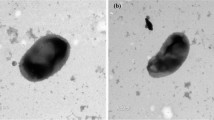
A novel Gram-stain-negative, aerobic and rod-shaped bacterium with a single polar flagellum or a stalk at the end of the cell, was isolated from maize roots in the Fangshan District of Beijing, People’s Republic of China. The new strain designated 774T produced indole acetic acid (IAA). The 16S rRNA gene sequence analysis indicated that strain 774T belongs to the genus Caulobacter and is closely related to Caulobacter flavus RHGG3T, Caulobacter zeae 410Tand Caulobacter radices 695T, all with sequence similarities of 99.9%. The genome size of strain774T was 5.4 Mb, comprising 5042 predicted genes with a DNA G+C content of 68.7%.Three striking lasso peptide biosynthetic gene clusters and two IAA synthesis genes belonging to the TPM pathway were also found in the genome of strain 774T. The average nucleotide identity values and digital DNA–DNA hybridization values of the strain774T with its closely phylogenetic neighbours were less than 91.5% and 45.0%, respectively, indicating a new Caulobacter species. The major fatty acids of strain774T were identified as C16: 0 (27.7%), summed feature 3 (C16: 1ω6c and/or C16: 1ω7c) (12.6%) and summed feature 8 (C18: 1ω7c and/or C18: 1ω6c) (42.9%).The major polar lipids consisted of phosphatidyl–glycerol and glycolipids. The predominant ubiquinone was identified as Quinone 10. Based on the polyphasic characterization, strain 774T represents a novel species of the genus Caulobacter, for which the name Caulobacter endophyticus sp. nov. is proposed with 774T (= CGMCC 1.16558T = DSM 106777T) as the type strain.


Similar content being viewed by others
References
Abraham WR, Strömpl C, Meyer H, Lindholst S, Moore RB, Christ R, Vancanneyt M, Tindall BJ, BennasarA TM (1999) Phylogeny and polyphasic taxonomy of Caulobacter species. Proposal of Maricaulis gen. nov.with Maricaulismaris (poindexter) comb. nov. as the type species, and emended description of the genera Brevundimonas and Caulobacter. Int J Syst Bacteriol 49:1053–1073
AbrahamWR MAJ, Lünsdorf H, Fischer R, Pawelczyk S, Vancanneyt M (2008) Phylogeny by a polyphasic approach of the order Caulobacterales, proposal of Caulobacter mirabilis sp.nov., Phenylobacterium haematophilum sp. nov. and Phenylobacterium conjunctum sp. nov., and emendation of the genus Phenylobacterium. Int J Syst Evol Microbiol 58:1939–1949
Bligh EG, Dyer JW (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) AntiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87
Bowers LE, Weaver RH, Grula EA, Edwards OF (1954) Studies on a strain of Caulobacter from water. I. Isolation and identification as Caulobacter vibrioides Henrici and Johnson with emended description. J Bacteriol 68:194–200
Breznak JA, Costilow RN (2007) Physicochemical factors in growth. Beveridge TJ, Breznak JA, Marzluf GA, Schmidt TM, Snyder LR(eds) Methods for general and molecular bacteriology, 3rd edn. American Society for Microbiology, Washington, pp 309–329
Chen H, Jogler M, Rohde M, Klenk HP, Busse HJ, Brian JT, Overmann J, Spröer C (2012) Reclassification and emended description of Caulobacter leidyi as Sphingomonasleidyi comb. nov., and emendation of the genus Sphingomonas. Int J Syst Evol Microbiol 62:2835–2843
Cherif H, Marasco R, Rolli E, Ferjani R, Fusi M, Soussi A, Mapelli F, Blilou I, Borin S, Boudabous A, Cherif A, Ouzari H (2015) Oasis desert farming selects environment-specific date palm root endophytic communities and cultivable bacteria that promote resistance to drought. Environ Microbiol Rep 7:668–678
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer S, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2,4-diaminobutyric acid. J Appl Microbiol 48:459–470
Delory GE, King EJ (1945) A sodium carbonate-bicarbonate buffer for alkaline phosphatases. Biochem J 39:245
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Gao M, Zhou JJ, Wang ET, Chen Q, Xu J, Sun JG (2015) Multiphasic characterization of a plant growth promoting bacterial strain, Burkholderia sp. 7016 and its effect on tomato growth in the field. J Integr Agric 14(9):1855–1863
Gao JL, Yuan M, Wang XM, Qiu TL, Lv FY, Sun JG (2016) Paenibacillus radicis sp. nov., a endophytic bacterium isolated from maize root in China. Int J Syst Evol Microbiol 66:801–811
Gao JL, Sun P, Wang XM, Lv FY, Mao XJ, Sun JG (2017) Rhizobium wenxiniae sp. nov. an endophytic bacterium isolated from maize root. Int J Syst Evol Microbiol 67:2798–2803
GaoJL SP, Sun XH, Tong S, Yan H, Sun JG (2018) Caulobacter zeae sp. nov. and Caulobacter radicis sp. nov., novel endophytic bacteria isolated from maize root (Zea mays L.). Syst Appl Microbiol 41:604–610
Gutierrez CK, Matsui GY, Lincoln DE, Lovell CR (2009) Production of the phytohormone indole-3-acetic acid by estuarine species of the genus Vibrio. Appl Environ Microbiol 75:2253–2258
Hegemann JD, Zimmermann M, Zhu S, Klug D, Marahiel MA (2013a) Lasso peptides from proteobacteria: Genome mining employing heterologous expression and mass spectrometry. Pept Sci 100:527–542
Hegemann JD, Zimmermann M, Xie X, Marahiel MA, M A, (2013b) Caulosegnins I-III: a highly diverse group of lasso peptides derived from a single biosynthetic gene cluster. J Am Chem Soc 135(1):210–222
Henrici AT, Johnson DE (1935) Studies of freshwater bacteria.II. Stalked bacteria, a new order of Schizomycetes. J Bacteriol 30:61–93
Jin L, Lee HG, Kim HS, Ahn CY, Oh HM (2013) Caulobacter daechungensis sp. nov., a stalked bacterium isolated from a eutrophic reservoir. Int J Syst Evol Microbiol 63:2559–2564
Jin L, La HJ, Lee HG, Lee JJ, Lee S, Oh HM (2014) Caulobacter profunda sp. nov., isolated from deep freshwater sediment. Int J Syst Evol Microbiol 64:762–767
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackerandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematic. Wiley, Chichester, pp 115–175
Li Y, Zirah S, Rebuffat S (2015) Lasso peptides: bacterial strategies to make and maintain bioactive entangled scaffolds. Springer, New York
Liu QM, Ten LN, Im WT, Lee ST, Yoon MH (2010) Caulobacter ginsengisoli sp. nov., a novel stalked bacterium isolated from ginseng cultivating soil. J Microbiol Biotechnol 20:15–20
Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1(1):1–6
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Minnikin D, O’Donnell A, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Appl Bacteriol 2:233–241
Montanez A, Blanco AR, Barlocco C, Beracochea M, Sicardi M (2012) Characterization of cultivable putative endophytic plant growth promoting bacteria associated with maize cultivars (zea mays l.) and their inoculation effects in vitro. Appl Soil Ecol 58:21–28
Moya G, Yan ZF, Won KH, Yang JE, Wang QJ, Kook MC, Yi TH (2017) Caulobacter hibisci sp. nov., isolated from rhizosphere of Hibiscus syriacus L. (Mugunghwa flower). Int J Syst Evol Microbiol 67:3167–3173
Poindexter JS (1964) Biological properties and classification of the Caulobacter group. Bacteriol Rev 28:231–295
Poindexter JS (2005) Genus, I. Caulobcter. In: Holt JG, Krieg NR, Sneath PH, Staley JT, Williams T (eds) Bergey’s manual of determinative bacteriology, 10th edn. The Williams & Wilkins Co, Baltimore, pp 287–303
Reasoner DJ, Geldreich EE (1985) A new medium for the enumeration and subculture of bacteria from potable water. Appl Environ Microbiol 49:1–7
Rolli E, MarascoR VG, Ettoumi B, Mapelli F, Deangelis ML, Gandolfi C, Casati E, Previtali F, Gerbino R, Cei FP, Borin S, Sorlini C, Daffonchio D (2015) Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ Microbiol 17:316–331
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101. Microbial ID Inc, Newark
Shao JH, Li SQ, Zhang N, Cui XS, Zhou X, Zhang GS, Zhang RF (2015) Analysis and cloning of the synthetic pathway of the phytohormone indole-3-acetic acid in the plant-beneficial Bacillus amyloliquefaciens SQR9. Microb Cell Fact 14:130
Sun LN, Yang ED, Wei JC, Tang XY, Cao YY (2015) Caulobacter flavus sp. nov. a stalked bacterium isolated from rhizosphere soil. Int J Syst Evol Microbiol 65:4374–4380
Sun LN, Yang ED, Hou XT, Wei JC, Yuan ZX, Wang WY (2017) Caulobacter rhizosphaerae sp. nov., a stalked bacterium isolated from rhizosphere soil. Int J Syst Evol Microbiol 67:1771–1776
Teale WD, Paponov IA, Palme K (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7:847–859
Urakami T, Oyanagi H, Araki H, Suzuki K, Komagata K (1990) Recharacterizati on and emended description of the genus Mycoplana and description of two new species, Mycoplana ramose and Mycoplanasegnis. Int J Syst Bacteriol 40:434–442
Wang Q, Zhu W, Wang ET, Zhang LS, Li X (2016) Genomic identification of rhizobia-related strains and threshold of ANI and core-genome for family, genus and species. Int J Environ Agri Res (IJOEAR) 2:76–86
Yang YY, Jin CZ, Jin FJ, Li TH, Lee JM, Kim CJ, Jin L (2020) Caulobacter soli sp. Nov. isolated from soil sampled at Jiri Mountain, Republic of Korea. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.004264
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhang P, Jin T, Sahu SK, Xu J, Shi Q, Liu H, Wang Y (2019) The distribution of tryptophan-dependent indole-3-acetic acid synthesis pathways in bacteria unraveled by large-scale genomic analysis. Molecules 24:1411
Zhao J, Zhao X, Wang J, Gong Q, Zhang XX, Zhang GS (2020) Isolation, identification and characterization of endophytic bacterium rhizobium oryzihabitans sp. nov, from rice root with biotechnological potential in agriculture. Microorganisms 8(4):608
Acknowledgements
This research work was supported financially by the National Natural Science Foundation of China (No. 31870003), Science & Technology Innovation Project of Beijing Academy of Agricultural and Forestry Sciences (KJCX20200426), Science & Technology Innovation Project of Beijing Academy of Agricultural and Forestry Sciences (KJCX20200110) and Beijing Postdoctoral Research Foundation (2020-ZZ-106).
Author information
Authors and Affiliations
Contributions
JGS and XHZ designed research and project outline. JLG, PBS, YCS, LWW and JX performed isolation, deposition and polyphasic taxonomy. PBS, GLW and YPD performed genome analysis. JLG and YCS drafted the manuscript. PBS, JGS and XHZ revised the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gao, Jl., Sun, P., Sun, Yc. et al. Caulobacter endophyticus sp. nov., an endophytic bacterium harboring three lasso peptide biosynthetic gene clusters and producing indoleacetic acid isolated from maize root. Antonie van Leeuwenhoek 114, 1213–1224 (2021). https://doi.org/10.1007/s10482-021-01593-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-021-01593-9