Abstract
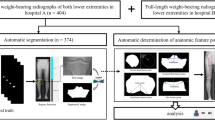
Kinematic tracking of native anatomy from stereo-radiography provides a quantitative basis for evaluating human movement. Conventional tracking procedures require significant manual effort and call for acquisition and annotation of subject-specific volumetric medical images. The current work introduces a framework for fully automatic tracking of native knee anatomy from dynamic stereo-radiography which forgoes reliance on volumetric scans. The method consists of three computational steps. First, captured radiographs are annotated with segmentation maps and anatomic landmarks using a convolutional neural network. Next, a non-convex polynomial optimization problem formulated from annotated landmarks is solved to acquire preliminary anatomy and pose estimates. Finally, a global optimization routine is performed for concurrent refinement of anatomy and pose. An objective function is maximized which quantifies similarities between masked radiographs and digitally reconstructed radiographs produced from statistical shape and intensity models. The proposed framework was evaluated against manually tracked trials comprising dynamic activities, and additional frames capturing a static knee phantom. Experiments revealed anatomic surface errors routinely below 1.0 mm in both evaluation cohorts. Median absolute errors of individual bone pose estimates were below 1.0\(^{\circ }\) or mm for 15 out of 18 degrees of freedom in both evaluation cohorts. Results indicate that accurate pose estimation of native anatomy from stereo-radiography may be performed with significantly reduced manual effort, and without reliance on volumetric scans.



Similar content being viewed by others
References
Hamilton, L. D., T. E. Andreassen, C. Myers, K. B. Shelburne, C. Clary, and P. J. Rullkoetter. Supine leg press as an alternative to standing lunge in high-speed stereo radiography. J. Biomech. 138:111118, 2022.
Myers, C. A., M. R. Torry, K. B. Shelburne, J. E. Giphart, R. F. LaPrade, S. L. Woo, et al. In vivo tibiofemoral kinematics during 4 functional tasks of increasing demand using biplane fluoroscopy. Am. J. Sports Med. 40(1):170–178, 2012.
Grood, E. S., and W. J. Suntay. A joint coordinate system for the clinical description of three-dimensional motions: application to the knee. J. Biomech. Eng. 105(2):136–144, 1983.
Tashman, S., D. Collon, K. Anderson, P. Kolowich, and W. Anderst. Abnormal rotational knee motion during running after anterior cruciate ligament reconstruction. Am. J. Sports Med. 32(4):975–983, 2004.
Martelli, S., N. Sancisi, M. Conconi, M. G. Pandy, M. E. Kersh, V. Parenti-Castelli, et al. The relationship between tibiofemoral geometry and musculoskeletal function during normal activity. Gait Posture. 80:374–382, 2020.
Navacchia, A., D. R. Hume, P. J. Rullkoetter, and K. B. Shelburne. A computationally efficient strategy to estimate muscle forces in a finite element musculoskeletal model of the lower limb. J. Biomech. 84:94–102, 2019.
Hamai, S., T. Moro-oka, H. Miura, T. Shimoto, H. Higaki, et al. Knee kinematics in medial osteoarthritis during in vivo weight-bearing activities. J. Orthop. Res. 27(12):1555–1561, 2009.
Kozanek, M., A. Hosseini, F. Liu, S. K. Van de Velde, T. J. Gill, H. E. Rubash, et al. Tibiofemoral kinematics and condylar motion during the stance phase of gait. J. Biomech. 42(12):1877–1884, 2009.
Leszko, F., K. R. Hovinga, A. L. Lerner, R. D. Komistek, and M. R. Mahfouz. In vivo normal knee kinematics: is ethnicity or gender an influencing factor? Clin. Orthop. Related Res. 469:95–106, 2011.
Qi, W., A. Hosseini, T. Y. Tsai, J. S. Li, H. E. Rubash, and G. Li. In vivo kinematics of the knee during weight bearing high flexion. J. Biomech. 46(9):1576–1582, 2013.
Seisler, A. R., and F. T. Sheehan. Normative three-dimensional patellofemoral and tibiofemoral kinematics: a dynamic, in vivo study. IEEE Trans. Biomed. Eng. 54(7):1333–1341, 2007.
Anderst, W., R. Zauel, J. Bishop, E. Demps, and S. Tashman. Validation of three-dimensional model-based tibio-femoral tracking during running. Med. Eng. Phys. 31(1):10–16, 2009.
Giphart, J. E., C. A. Zirker, C. A. Myers, W. W. Pennington, and R. F. LaPrade. Accuracy of a contour-based biplane fluoroscopy technique for tracking knee joint kinematics of different speeds. J. Biomech. 45(16):2935–2938, 2012.
Ivester, J. C., A. J. Cyr, M. D. Harris, M. J. Kulis, P. J. Rullkoetter, and K. B. Shelburne. A reconfigurable high-speed stereo-radiography system for sub-millimeter measurement of in vivo joint kinematics. J. Med. Device. 9(4):041009, 2015.
Fregly, B. J., H. A. Rahman, and S. A. Banks. Theoretical accuracy of model-based shape matching for measuring natural knee kinematics with single-plane fluoroscopy. J. Biomech. Eng. 127(4):692–699, 2005.
Postolka, B., R. List, B. Thelen, P. Schütz, W. R. Taylor, and G. Zheng. Evaluation of an intensity-based algorithm for 2D/3D registration of natural knee videofluoroscopy data. Med. Eng. Phys. 77:107–113, 2020.
You, B. M., P. Siy, W. Anderst, and S. Tashman. In vivo measurement of 3-D skeletal kinematics from sequences of biplane radiographs: application to knee kinematics. IEEE Trans. Med. Imaging. 20(6):514–525, 2001.
Asano, T., M. Akagi, K. Tanaka, J. Tamura, and T. Nakamura. In vivo three-dimensional knee kinematics using a biplanar image-matching technique. Clin. Orthop. Related Res. 388:157–166, 2001.
Hamilton, L. D., T. E. Andreassen, C. A. Myers, K. B. Shelburne, C. W. Clary, and P. J. Rullkoetter. Knee pivot location in asymptomatic older adults. J. Biomech. 149:111487, 2023.
Kefala, V., A. J. Cyr, M. D. Harris, D. R. Hume, B. S. Davidson, R. H. Kim, et al. Assessment of knee kinematics in older adults using high-speed stereo radiography. Med. Sci. Sports Exercise. 49(11):2260, 2017.
Akbari-Shandiz, M., R. L. Lawrence, A. M. Ellingson, C. P. Johnson, K. D. Zhao, and P. M. Ludewig. MRI vs CT-based 2D–3D auto-registration accuracy for quantifying shoulder motion using biplane video-radiography. J. Biomech. 82:375–380, 2019.
Englander, Z. A., J. T. Martin, P. K. Ganapathy, W. E. Garrett, and L. E. DeFrate. Automatic registration of MRI-based joint models to high-speed biplanar radiographs for precise quantification of in vivo anterior cruciate ligament deformation during gait. J. Biomech. 81:36–44, 2018.
Ku, P. C., A. Martin-Gomez, C. Gao, R. Grupp, S. C. Mears, and M. Armand. Towards 2D/3D registration of the preoperative MRI to intraoperative fluoroscopic images for visualisation of bone defects. Comput. Methods Biomech. Biomed. Eng. Imaging. Vis. 11(4):1096–1105, 2023.
Williams, D. E. Development and Validation of a Biplane Fluoroscopy System to Quantify In-Vivo Knee Kinematics. Cardiff: Cardiff University, 2018.
Ehlke, M., H. Ramm, H. Lamecker, H. C. Hege, and S. Zachow. Fast generation of virtual X-ray images for reconstruction of 3D anatomy. IEEE Trans. Vis. Comput. Gr. 19(12):2673–2682, 2013.
Jacobs, F., E. Sundermann, B. De Sutter, M. Christiaens, and I. Lemahieu. A fast algorithm to calculate the exact radiological path through a pixel or voxel space. J. Comput. Info. Technol. 6(1):89–94, 1998.
Siddon, R. L. Fast calculation of the exact radiological path for a three-dimensional CT array. Med. Phys. 12(2):252–255, 1985.
Burton, W., I. R. Crespo, T. Andreassen, M. Pryhoda, A. Jensen, C. Myers, et al. Fully automatic tracking of native glenohumeral kinematics from stereo-radiography. Comput. Biol. Med. 107189, 2023.
Burton, W. S., C. A. Myers, A. Jensen, L. Hamilton, K. B. Shelburne, S. A. Banks, et al. Automatic tracking of healthy joint kinematics from stereo-radiography sequences. Comput. Biol. Med. 139:104945, 2021.
Grupp, R. B., M. Unberath, C. Gao, R. A. Hegeman, R. J. Murphy, C. P. Alexander, et al. Automatic annotation of hip anatomy in fluoroscopy for robust and efficient 2D/3D registration. Int. J. Comput. Assist. Radiol. Surg. 15:759–769, 2020.
Liao, H., W. A. Lin, J. Zhang, J. Zhang, J. Luo, and S. K. Zhou. Multiview 2D/3D rigid registration via a point-of-interest network for tracking and triangulation. Proc. IEEE/CVF Conf. Comput. Vision Pattern Recognit. 12638–12647, 2019.
Gu, W., C. Gao, R. Grupp, J. Fotouhi, and M. Unberath. Extended capture range of rigid 2D/3D registration by estimating Riemannian pose gradients. In: Machine Learning in Medical Imaging: 11th International Workshop, MLMI 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, October 4, 2020, Proceedings 11. Springer, 2020, p. 281–291.
Banks, S. A., and W. A. Hodge. Accurate measurement of three-dimensional knee replacement kinematics using single-plane fluoroscopy. IEEE Trans. Biomed. Eng. 43(6):638–649, 1996.
Jensen, A. J., P. D. Flood, L. S. Palm-Vlasak, W. S. Burton, A. Chevalier, P. J. Rullkoetter, et al. Joint track machine learning: an autonomous method of measuring total knee arthroplasty kinematics from single-plane X-ray images. J. Arthroplast. 2023.
Miao, S., R. Liao, J. Lucas, and C. Chefd’hotel. Toward accurate and robust 2-D/3-D registration of implant models to single-plane fluoroscopy. In: International Workshop on Medical Imaging and Virtual Reality. Springer, 2013, pp. 97–106.
Varnavas, A., T. Carrell, and G. Penney. Fully automated initialisation of 2D-3D image registration. In: IEEE 10th International Symposium on Biomedical Imaging. IEEE, 2013, pp. 568–571.
Abumoussa, A., V. Gopalakrishnan, B. Succop, M. Galgano, S. Jaikumar, Y. Z. Lee, et al. Machine learning for automated and real-time two-dimensional to three-dimensional registration of the spine using a single radiograph. Neurosurg. Focus. 54(6):E16, 2023.
Esteban, J., M. Grimm, M. Unberath, G. Zahnd, and N. Navab. Towards fully automatic X-ray to CT registration. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part VI 22. Springer, 2019, pp. 631–639.
Flood, P. D., and S. A. Banks. Automated registration of 3-D knee implant models to fluoroscopic images using Lipschitzian optimization. IEEE Trans. Med. Imaging. 37(1):326–335, 2017.
Gao, C., X. Liu, W. Gu, B. Killeen, M. Armand, R. Taylor, et al. Generalizing spatial transformers to projective geometry with applications to 2D/3D registration. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part III 23. Springer, 2020, pp. 329–339.
Gao, C., A. Feng, X. Liu, R. H. Taylor, M. Armand, and M. Unberath. A fully differentiable framework for 2D/3D registration and the projective spatial transformers. IEEE Trans. Med. Imaging. 43(1):275–285, 2023.
Gopalakrishnan, V., and P. Golland. Fast auto-differentiable digitally reconstructed radiographs for solving inverse problems in intraoperative imaging. In: Workshop on Clinical Image-Based Procedures. Springer, 2022, pp. 1–11.
Mahfouz, M. R., W. A. Hoff, R. D. Komistek, and D. A. Dennis. A robust method for registration of three-dimensional knee implant models to two-dimensional fluoroscopy images. IEEE Trans. Med. Imaging. 22(12):1561–1574, 2003.
Mu, S. Skeletal Kinematic Measurement Using Model-Image Registration and Mechanical Constraints. Florida: University of Florida, 2010.
Otake, Y., M. Armand, R. S. Armiger, M. D. Kutzer, E. Basafa, P. Kazanzides, et al. Intraoperative image-based multiview 2D/3D registration for image-guided orthopaedic surgery: incorporation of fiducial-based C-arm tracking and GPU-acceleration. IEEE Trans. Med. Imaging. 31(4):948–962, 2011.
Shetty, K., A. Birkhold, N. Strobel, B. Egger, S. Jaganathan, M. Kowarschik, et al. Deep learning compatible differentiable X-ray projections for inverse rendering. In: Bildverarbeitung für die Medizin 2021: Proceedings, German Workshop on Medical Image Computing, Regensburg, March 7-9, Springer, 2021. pp. 290–295.
Van Der Bom, I., S. Klein, M. Staring, R. Homan, L. W. Bartels, and J. P. Pluim. Evaluation of optimization methods for intensity-based 2D-3D registration in X-ray guided interventions. In: Medical Imaging 2011: Image Processing. vol. 7962. SPIE, 2021, pp. 657–671.
Varnavas, A., T. Carrell, and G. Penney. Fully automated 2D–3D registration and verification. Med. Image Anal. 26(1):108–119, 2015.
Chou, C. R., B. Frederick, G. Mageras, S. Chang, and S. Pizer. 2D/3D image registration using regression learning. Comput. Vision Image Underst. 117(9):1095–1106, 2013.
Gouveia, A. R., C. Metz, L. Freire, P. Almeida, and S. Klein. Registration-by-regression of coronary CTA and X-ray angiography. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 5(3):208–220, 2017.
Miao, S., Z. J. Wang, and R. Liao. A CNN regression approach for real-time 2D/3D registration. IEEE Trans. Med. Imaging. 35(5):1352–1363, 2016.
Xie, Y., C. Meng, S. Guan, and Q. Wang. Single shot 2D3D image regisraton. In: 10th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI). IEEE, 2017, pp. 1–5.
Zheng, J., S. Miao, and R. Liao. Learning CNNS with pairwise domain adaption for real-time 6-DoF ultrasound transducer detection and tracking from X-ray images. In: Medical Image Computing and Computer-Assisted Intervention- MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, September 11-13, 2017, Proceedings, Part II 20. Springer, 2017, pp. 646–654.
Geng, H., D. Xiao, S. Yang, J. Fan, T. Fu, Y. Lin, et al. CT2X-IRA: CT to X-ray image registration agent using domain-cross multi-scale-stride deep reinforcement learning. Phys. Med. Biol. 68(17):175024, 2023.
Hu, J., Z. Luo, X. Wang, S. Sun, Y. Yin, K. Cao, et al. End-to-end multimodal image registration via reinforcement learning. Med. Image Anal. 68:101878, 2021.
Liao, R., S. Miao, P. de Tournemire, S. Grbic, A. Kamen, T. Mansi, et al. An artificial agent for robust image registration. Proc. AAAI Conf. Artif. Intell. 2017.
Miao, S., S. Piat, P. Fischer, A. Tuysuzoglu, P. Mewes, T. Mansi, et al. Dilated FCN for multi-agent 2D/3D medical image registration. Proc. AAAI Conf. Artif. Intell. 2018.
Toth, D., S. Miao, T. Kurzendorfer, C. A. Rinaldi, R. Liao, T. Mansi, et al. 3D/2D model-to-image registration by imitation learning for cardiac procedures. Int. J. Comput. Assist. Radiol. Surg. 13:1141–1149, 2018.
Jaganathan, S., J. Wang, A. Borsdorf, and A. Maier. Learning the update operator for 2D/3D image registration. In: Bildverarbeitung für die Medizin 2021: Proceedings, German Workshop on Medical Image Computing, Regensburg, March 7-9, 2021. Springer, 2021, pp. 117–122.
Jaganathan, S., M. Kukla, J. Wang, K. Shetty, and A. Maier. Self-supervised 2D/3D registration for X-ray to CT image fusion. Proc. IEEE/CVF Winter Conf. Appl. Comput. Vis. 2788–2798, 2023.
Schaffert, R., M. Weiß, J. Wang, A. Borsdorf, and A.Maier. Learning-based correspondence estimation for 2-D/3-D registration. In: Bildverarbeitung für die Medizin 2020: Algorithmen– Systeme–Anwendungen. Proceedings des Workshops vom 15. bis 17. März 2020 in Berlin. Springer, 2020, pp. 222–228.
Schaffert, R., J. Wang, P. Fischer, A. Borsdorf, and A. Maier. Learning an attention model for robust 2-D/3-D registration using point-to-plane correspondences. IEEE Trans. Med. Imaging. 39(10):3159–3174, 2020.
Burton, W., C. Myers, K. Shelburne, M. Stefanovic, and P. Rullkoetter. Fully automatic tracking of native knee kinematics from stereo-radiography with digitally reconstructed radiographs. J. Biomech. 163:107189, 2023.
Reyneke, C. J. F., M. Lüthi, V. Burdin, T. S. Douglas, T. Vetter, and T. E. Mutsvangwa. Review of 2-D/3-D reconstruction using statistical shape and intensity models and X-ray image synthesis: toward a unified framework. IEEE Rev. Biomed. Eng. 12:269–286, 2018.
Chênes, C., and J. Schmid. Revisiting contour-driven and knowledge-based deformable models: application to 2D-3D proximal femur reconstruction from X-ray images. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part VI 24. Springer, 2021, pp. 451–460.
Kurazume, R., K. Nakamura, T. Okada, Y. Sato, N. Sugano, T. Koyama, et al. 3D reconstruction of a femoral shape using a parametric model and two 2D fluoroscopic images. Comput. Vision Image Underst. 113(2):202–211, 2009.
Väänänen, S. P., L. Grassi, G. Flivik, J. S. Jurvelin, and H. Isaksson. Generation of 3D shape, density, cortical thickness and finite element mesh of proximal femur from a DXA image. Med. Image Anal. 24(1):125–134, 2015.
Yao, J. A Statistical Bone Density Atlas and Deformable Medical Image Registration. Baltimore: The Johns Hopkins University, 2002.
Zhu, Z., and G. Li. Construction of 3D human distal femoral surface models using a 3D statistical deformable model. J. Biomech. 44(13):2362–2368, 2011.
Baka, N., B. L. Kaptein, M. de Bruijne, T. van Walsum, J. Giphart, W. J. Niessen, et al. 2D–3D shape reconstruction of the distal femur from stereo X-ray imaging using statistical shape models. Med. Image Anal. 15(6):840–850, 2011.
Baka, N., B. L. Kaptein, J. E. Giphart, M. Staring, M. de Bruijne, B. P. Lelieveldt, et al. Evaluation of automated statistical shape model based knee kinematics from biplane fluoroscopy. J Biomech. 47(1):122–129, 2014.
Li, J. S., T. Y. Tsai, S. Wang, P. Li, Y. M. Kwon, A. Freiberg, et al. Prediction of in vivo knee joint kinematics using a combined dual fluoroscopy imaging and statistical shape modeling technique. J. Biomech. Eng. 136(12):124503, 2014.
Lu, H. Y., C. C. Lin, K. S. Shih, T. W. Lu, M. Y. Kuo, S. Y. Li, et al. Integration of statistical shape modeling and alternating interpolation-based model tracking technique for measuring knee kinematics in vivo using clinical interleaved bi-plane fluoroscopy. PeerJ. 11:e15371, 2023.
Smoger, L. M., K. B. Shelburne, A. J. Cyr, P. J. Rullkoetter, and P. J. Laz. Statistical shape modeling predicts patellar bone geometry to enable stereo-radiographic kinematic tracking. J. Biomech. 58:187–194, 2017.
Valenti, M., E. De Momi, W. Yu, G. Ferrigno, M. Akbari Shandiz, C. Anglin, et al. Fluoroscopy-based tracking of femoral kinematics with statistical shape models. Int. J. Comput. Assist. Radiol. Surg. 11:757–765, 2016.
Gaffney, B. M., T. J. Hillen, J. J. Nepple, J. C. Clohisy, and M. D. Harris. Statistical shape modeling of femur shape variability in female patients with hip dysplasia. J. Orthop. Res. 37(3):665–673, 2019.
Sintini, I., W. S. Burton, P. Sade Sr., J. M. Chavarria, and P. J. Laz. Investigating gender and ethnicity differences in proximal humeral morphology using a statistical shape model. J. Orthop. Res. 36(11):3043–3052, 2018.
Bryan, R., P. S. Mohan, A. Hopkins, F. Galloway, M. Taylor, and P. B. Nair. Statistical modelling of the whole human femur incorporating geometric and material properties. Med. Eng. Phys. 32(1):57–65, 2010.
Burton, W. S., I. Sintini, J. M. Chavarria, J. R. Brownhill, and P. J. Laz. Assessment of scapular morphology and bone quality with statistical models. Comput. Methods Biomech. Biomed. Eng. 22(4):341–351, 2019.
Yu, W., M. Tannast, and G. Zheng. Non-rigid free-form 2D–3D registration using a B-spline-based statistical deformation model. Pattern Recognit. 63:689–699, 2017.
Zheng, G., and W. Yu. Statistical shape and deformation models based 2D–3D reconstruction, Chapter 12. In: Statistical Shape and Deformation Analysis, Amsterdam: Elsevier, 2017, pp. 329–349.
Maken, P., and A. Gupta. 2D-to-3D: a review for computational 3D image reconstruction from X-ray images. Arch. Comput. Methods Eng. 30(1):85–114, 2023.
Hampali, S. 3D Shape Reconstruction of Knee Bones from Low Radiation X-ray Images Using Deep Learning.
Sun, W., Y. Zhao, J. Liu, and G. Zheng. LatentPCN: latent space-constrained point cloud network for reconstruction of 3D patient-specific bone surface models from calibrated biplanar X-ray images. Int. J. Comput. Assist. Radiol. Surg. 1–11, 2023.
Bayat, A., A. Sekuboyina, J. C. Paetzold, C. Payer, D. Stern, M. Urschler, et al. Inferring the 3D standing spine posture from 2D radiographs. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2020: 23rd International Conference, Lima, Peru, October 4–8, 2020, Proceedings, Part VI 23. Springer, 2020, pp. 775–784.
Chen, C. C., and Y. H. Fang. Using bi-planar X-ray images to reconstruct the spine structure by the convolution neural network. In: Future Trends in Biomedical and Health Informatics and Cybersecurity in Medical Devices: Proceedings of the International Conference on Biomedical and Health Informatics, ICBHI 2019, 17-20 April 2019, Taipei, Taiwan. Springer, 2020, pp. 80–85.
Kasten, Y., D. Doktofsky, and I. Kovler. End-to-end convolutional neural network for 3D reconstruction of knee bones from bi-planar X-ray images. In: Machine Learning for Medical Image Reconstruction: Third International Workshop, MLMIR 2020, Held in Conjunction with MICCAI 2020, Lima, Peru, October 8, 2020, Proceedings 3. Springer, 2020, pp. 123–133.
Shiode, R., M. Kabashima, Y. Hiasa, K. Oka, T. Murase, Y. Sato, et al. 2D–3D reconstruction of distal forearm bone from actual X-ray images of the wrist using convolutional neural networks. Sci. Rep. 11(1):15249, 2021.
Almeida, D. F., P. Astudillo, and D. Vandermeulen. Three-dimensional image volumes from two-dimensional digitally reconstructed radiographs: a deep learning approach in lower limb CT scans. Med. Phys. 48(5):2448–2457, 2021.
Henzler, P., V. Rasche, T. Ropinski, and T. Ritschel. Single-image tomography: 3D volumes from 2D cranial X-rays. Comp. Graph. Forum. 37(2):377–388, 2018.
Shen, L., W. Zhao, and L. Xing. Patient-specific reconstruction of volumetric computed tomography images from a single projection view via deep learning. Nat. Biomed. Eng. 3(11):880–888, 2019.
Ying, X., H. Guo, K. Ma, J. Wu, Z. Weng, and Y. Zheng. X2CT-GAN: reconstructing CT from biplanar X-rays with generative adversarial networks. Proc. IEEE/CVF Conf. Comput. Vision Pattern Recognit. 10619–10628, 2019.
Bayat, A., D. F. Pace, A. Sekuboyina, C. Payer, D. Stern, M. Urschler, et al. Anatomy-aware inference of the 3D standing spine posture from 2D radiographs. Tomography. 8(1):479–496, 2022.
Nakao, M., F. Tong, M. Nakamura, and T. Matsuda. Image-to-graph convolutional network for deformable shape reconstruction from a single projection image. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part IV 24. Springer, 2010 pp. 259–268.
Van Houtte, J., X. Gao, J. Sijbers, and G. Zheng. 2D/3D registration with a statistical deformation model prior using deep learning. In: IEEE EMBS International Conference on Biomedical and Health Informatics (BHI). IEEE, 2021, pp. 1–4.
Van Houtte, J., E. Audenaert, G. Zheng, and J. Sijbers. Deep learning-based 2D/3D registration of an atlas to biplanar X-ray images. Int. J. Comput. Assist. Radiol. Surg. 17(7):1333–1342, 2022.
Lasserre, J. B. Moments, positive polynomials and their applications. Singapore: World Scientific, 2009.
Khoo, Y., and A. Kapoor. Non-iterative rigid 2D/3D point-set registration using semidefinite programming. IEEE Trans. Image Process. 25(7):2956–2970, 2016.
Hansen, N., and A. Auger. CMA-ES: evolution strategies and covariance matrix adaptation. Proc. 13th Ann. Conf. Companion Genet. Evol. Comput. 991–1010, 2011.
Brainerd, E. L., D. B. Baier, S. M. Gatesy, T. L. Hedrick, K. A. Metzger, S. L. Gilbert, et al. X-ray reconstruction of moving morphology (XROMM): precision, accuracy and applications in comparative biomechanics research. J. Exp. Zool. A Ecol. Genet. Physiol. 313(5):262–279, 2010.
Knörlein, B. J., D. B. Baier, S. M. Gatesy, J. Laurence-Chasen, and E. L. Brainerd. Validation of XMALab software for marker-based XROMM. J. Exp. Biol. 219(23):3701–3711, 2016.
Paszke, A., S. Gross, S. Chintala, G. Chanan, E. Yang, Z. DeVito, et al. Automatic differentiation in PyTorch. 2017.
Lofberg, J. YALMIP: A toolbox for modeling and optimization in MATLAB. In: IEEE International Conference on Robotics and Automation (IEEE Cat. No. 04CH37508). IEEE, 2004, pp. 284–289.
ApS, M. Mosek optimization toolbox for MATLAB user’s guide and reference manual, version 4:1, 2019.
Klöckner, A., N. Pinto, Y. Lee, B. Catanzaro, P. Ivanov, and A. Fasih. PyCUDA and PyOpenCL: a scripting-based approach to GPU run-time code generation. Parallel Comput. 38(3):157–174, 2012.
Besl, P. J., and N. D. McKay. Method for registration of 3-D shapes. In: Sensor fusion IV: control paradigms and data structures. vol. 1611. SPIE, 1992, pp. 586–6062.
Dagneaux, L., P. Thoreux, B. Eustache, F. Canovas, and W. Skalli. Sequential 3D analysis of patellofemoral kinematics from biplanar X-rays: In vitro validation protocol. Orthop. Traumatol. Surg. Res. 101(7):811–818, 2015.
Langlois, K., H. Pillet, F. Lavaste, G. Rochcongar, P. Rouch, P. Thoreux, et al. 3D sequential kinematics of the femoro-tibial joint of normal knee from multiple bi-planar X-rays: accuracy and repeatability. IRBM. 39(4):251–260, 2018.
Burton, W., C. Myers, C. Clary, C. Fitzpatrick, and P. Rullkoetter. Automated patient-specific simulation of implanted THA mechanics from CT. In: Orthopaedic Research Society. 2023.
Funding
This work was funded by the University of Denver.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Associate Editor Joel Stitzel oversaw review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Burton, W., Myers, C., Stefanovic, M. et al. Scan-Free and Fully Automatic Tracking of Native Knee Anatomy from Dynamic Stereo-Radiography with Statistical Shape and Intensity Models. Ann Biomed Eng (2024). https://doi.org/10.1007/s10439-024-03473-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10439-024-03473-5