Abstract
Classifying, describing and understanding the natural environment is an important element of studies of human, animal and ecosystem health, and baseline ecological data are commonly lacking in remote environments of the world. Human African trypanosomiasis is an important constraint on human well-being in sub-Saharan Africa, and spillover transmission occurs from the reservoir community of wild mammals. Here we use robust and repeatable methodology to generate baseline datasets on vegetation and mammal density to investigate the ecology of warthogs (Phacochoerus africanus) in the remote Luambe National Park in Zambia, in order to further our understanding of their interactions with tsetse (Glossina spp.) vectors of trypanosomiasis. Fuzzy set theory is used to produce an accurate landcover classification, and distance sampling techniques are applied to obtain species and habitat level density estimates for the most abundant wild mammals. The density of warthog burrows is also estimated and their spatial distribution mapped. The datasets generated provide an accurate baseline to further ecological and epidemiological understanding of disease systems such as trypanosomiasis. This study provides a reliable framework for ecological monitoring of wild mammal densities and vegetation composition in remote, relatively inaccessible environments.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding the structure of natural ecosystems forms the basis for understanding the processes within those ecosystems, including the transmission of infectious and vector-borne diseases. Remotely sensed datasets and geographical information systems (GIS) have been widely used to further our understanding of these systems. This technology has not only helped in the study of the global drivers of ecological change, but is also invaluable for understanding the biotic and abiotic factors influencing ecosystems at much smaller scales. GIS technology has become an integral component of many conservation programmes and the development of trans-disciplinary approaches such as Conservation Medicine, One Health and EcoHealth have highlighted its utility. However, recent moves to adopt ecosystem-based approaches within conservation and development programmes have highlighted frequent deficiencies in baseline ecological data, particularly in developing countries (Rapport et al. 1998).
For an area with such an internationally acclaimed biodiversity, relatively little ecological data exist for the Luangwa Valley in eastern Zambia (latitude −10.4° to −15.6° and longitude 30.2° to 33.1°). Although many valuable mapping and vegetation studies have been conducted, they either lack detail (Trapnell 1950; Naylor et al. 1973; Phiri 1989; Marks 2005) or have restricted geographical coverage (Astle et al. 1969; Smith 1998; Yang and Prince 2000). Similarly, published faunal surveys for the Luangwa Valley are rare and no peer-reviewed published data are available for many areas. Studies have been conducted in game management areas (GMAs) surrounding some of the national parks (Ndhlovu and Balakrishnan 1991; Lewis et al. 2011), and many of the species recorded historically in the mid-Luangwa Valley have been documented (Astle 1999). Aerial surveys have been conducted in the core parts of the Luangwa Valley on behalf of the Zambian Wildlife Authority (ZAWA) (Simukonda 2011) and as part of the Community Markets for Conservation Programme (COMACO) (Olive et al. 2012; Frederick 2013). The population of hippopotamus (Hippopotamus amphibious) has recently been surveyed and extensively studied (Wilbroad and Milanzi 2010; Chansa et al. 2011a; Chansa et al. 2011b). However, there is a clear need for more high-resolution data to enable active monitoring of ecosystem health in the valley.
There has been much interest in the role of warthogs (Phacochoerus africanus) as natural reservoir hosts for African swine fever (Plowright et al. 1969; Wilkinson et al. 1988), trypanosomiasis (Dillmann and Townsend 1979; Claxton et al. 1992) and bovine tuberculosis (Bengis et al. 2002; Michel et al. 2006). Warthog burrows not only provide a refuge for warthogs from predators and extremes of temperature, but they also provide a refuge for many parasites (Cumming 1975; Somers et al. 1994). The cool shady conditions in the entrance to warthog burrows provide an ideal refuge for tsetse flies during the heat of the day (Pilson and Pilson 1967), and the burrows are important sites for larviposition by female flies (Leak 1998). Warthogs are also a preferred host for Glossina morsitans species of tsetse flies, and a close ecological association between tsetse and warthog has been proposed (Pilson and Pilson 1967; Torr 1994; Leak 1998). A study of tsetse ecology in Luambe National Park (LNP) revealed that Combretum-Terminalia vegetation supports the highest apparent density of G. m. morsitans and thicket the highest apparent density of G. pallidipes (Anderson 2009). Warthogs have been shown to carry a moderate prevalence of trypanosomes and the human-infective Trypanosoma brucei rhodesiense, the cause of human African trypanosomiasis (HAT), has been identified in warthogs in the Luangwa Valley (Dillmann and Townsend 1979; Anderson et al. 2011). As a wide variety of other hosts are fed on by tsetse to varying degrees (Clausen et al. 1998) and are components of the natural reservoir community for trypanosomiasis in the Luangwa Valley (Anderson et al. 2011), it is important to understand more about the density and distribution of wild animal hosts within these ecosystems.
The majority of investigations into trypanosomiasis in wildlife have focussed on estimation of the prevalence of infection. Historically, prevalence was largely interpreted in terms of host susceptibility to infection and, to a lesser extent, host preference by tsetse. However, the importance of ecological and behavioural factors in the transmission of wildlife disease is now recognised (Cross et al. 2009). Factors such as habitat preference, resource use, territoriality, group size and group density contribute to a complex social and spatial structure for wildlife disease. Understanding the structure and distribution of both plant and animal communities is therefore critical for clarifying the nature of contact between hosts and vectors, and its impact on disease transmission. In a detailed review of the ecological factors influencing the epidemiology of trypanosomiasis in the Luangwa and Zambezi Valley ecosystems, Munang’andu et al. (2012) identified host distribution and abundance as having a significant influence on the survival of tsetse and therefore on trypanosomiasis epidemiology. Many other factors are also important including daily activity patterns of hosts and seasonal migration behaviour. Tsetse distribution and abundance is largely driven by climatic factors, host abundance and vegetation (Robinson et al. 1997). A better understanding of the distribution and characteristics of both mammal and plant communities is therefore likely to improve our management of HAT.
Here we generate accurate high-resolution datasets of vegetation and large mammal density in the remote, relatively inaccessible LNP within the Luangwa Valley, in order to investigate the ecology of warthogs and to further our understanding of their interactions with Glossina spp., vectors of trypanosomiasis.
Methods
Study Area
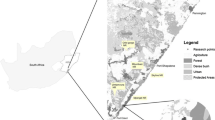
The Luangwa Valley lies in Muchinga, Eastern and Central Provinces of Zambia, forming an extension of the Great Rift Valley. LNP is a relatively small national park in the mid-Luangwa Valley, situated between the larger North and South Luangwa national parks on the other side of the Luangwa River (Fig. 1). It is poorly developed with minimal infrastructure, few roads and little accessibility during the rains (December to March). It is situated close to the historical sleeping sickness nidus in Nabwalia (Kinghorn et al. 1913).
Landcover Classification
Landsat 7 ETM+ Data
Landsat 7 ETM+ data with a spatial resolution of 30 m were selected for this study. The most recent L1G Landsat satellite image covering the study area was downloaded from the Global Land Cover Facility maintained by the University of Maryland (image acquisition date 04/10/2001, path 170, row 069, cloud cover 0%) (NASA Landsat Programme 2004). Proprietary satellite image processing software Erdas Imagine 8.4 (Leica Geosystems AG, Atlanta, USA) was used for all image processing and classification procedures.
Classification Scheme
The classification scheme selected is, to a degree, dictated by the objectives of the study in question (Congalton 1991). In this case, an important objective was to produce a dataset suitable for use as a GIS base layer for the design and spatial analysis of warthog and tsetse surveys planned for the park. Therefore, the classification level needed to distinguish between woodland classes, for example, rather than to simply classify to the broad physiognomic vegetation unit level (i.e. woodland).Footnote 1 The classification of vegetation at the physiognomic level by White (1983) was first used to understand the vegetation units represented in the park. These physiognomic units were then further divided into individual land cover classes (Table 1). Particular reference was made to the previous detailed studies of the vegetation by Astle et al. (1969) and Smith (1998) with allowances made for local differences in vegetation type found in LNP.
Ground-Truthing
Ground-truthing was conducted to confirm the classification scheme and collect reference training and test data in August and September, 2005. Evaluation of vegetation class relied on qualitative observations of vegetation physiognomy and predominant tree or shrub species present. Tree and plant species were identified with assistance from standard field guides covering the southern African region (Van Wyk and Van Wyk 1997; Coates Palgrave 2003a) and the Luangwa Valley (Smith 1995; Coates Palgrave 2003b). Spatial area was used to create a sample frame for collection of reference data, with polygons of homogenous vegetation as the sample unit. Twenty or more sample units were collected for all classes using a hand-held global positioning system (GPS), except for acacia woodland (sixteen) and hill scrub miombo woodland (nine) whose limited distribution precluded the collection of more data. For practical reasons, reference data for the water class was created from the non-classified Landsat image.
Classification
As collection of truly homogeneous polygons of vegetation was difficult due to the occurrence of natural mosaics, a classification approach based on fuzzy set theory was used (Wang 1990; Foody 1996). Fuzzy set theory allows for degrees of truth to be represented in algorithms allowing for joint membership of sets, or fuzzy boundaries. In image classification, it may be used to replace conventional probability theory in the classification process to create a fuzzy partition of the spectral space. This allows joint membership of classes by pixels represented by membership grades, the important feature being that mixed pixels are represented in the output classified image.
Reference polygons were firstly converted into regularly spaced points at 30-m intervals (each point effectively representing the value of one pixel) as the accuracy assessment algorithm required point data rather than polygon data. A subset of 25% of the points in each class were randomly selected and withheld from the training data, to be used as the test data. The training data were then used to create spectral signatures for each vegetation class. As the reflectance and emittance properties vary for different vegetation types, a spectral response pattern referred to as a spectral signature may be produced for each class from the training data (Lillesand et al. 2004). The image was classified using a maximum likelihood classifier with the additional activation of the fuzzy classification function (Pouncey et al. 1999). The feature space non-parametric decision rule was applied first and pixels in areas of overlap using this algorithm were then classified using the maximum likelihood parametric decision rule. Any unclassified pixels using the non-parametric decision rule were also classified using the parametric maximum likelihood rule. The option to select eight best classes was chosen resulting in an eight-layered fuzzy image. This was then processed into one layer to produce an output land cover map using the fuzzy convolution facility (Pouncey et al. 1999). A 3 × 3 window size was selected and the neighbour weighting option used with a neighbourhood weight factor of 0.5. All eight layers were used to perform the operation.
Image Evaluation
Classified images were examined by direct visualisation of the output image and graphical evaluation of spectral response patterns. Transformed divergence values were calculated to assess signature separability. An error matrix was generated to compare test and classified data; the overall accuracy, overall kappa statistic and modified estimator of kappa for stratified random sampling were calculated (Stehman 1997). Class level producer’s and user’s accuracies were calculated along with the conditional kappa values. The accuracy of the classification produced by the fuzzy algorithm was compared to a standard maximum likelihood classification using McNemar’s test for significance with the continuity correction (Foody 2004).
Distance Sampling Survey
Transect Design
The newly created land cover dataset was used to design a mammal density survey using distance sampling methods. The primary objective was to estimate the abundance of warthog and other mammalian hosts of trypanosomes and the secondary objective to map warthog burrow distribution. A ground transect survey was used rather than aerial survey techniques to enable more accurate estimation of the density of smaller species which are important hosts for trypanosomiasis. The study was conducted during the dry season from August to October, 2006, in the north of the park (the location for a concurrent tsetse survey). A single random starting point was generated to create a systematic grid of 40 parallel transects perpendicular to the Luangwa River. The length of each transect was 4.5 km and the distance between parallel transects was 250 m, providing a total survey length of 180 km (inset map, Fig. 1).
Survey Protocol
The same personnel were used throughout the survey, one being an observer and one a measurer and recorder. Transects were conducted on foot using a hand-held GPS and all observations of mammal species and warthog burrows were recorded. The perpendicular distance from the transect line was measured using a laser range-finder to ensure accuracy. All transects were started at approximately 6:30 am, the peak activity period for the majority of species of interest, to ensure consistency across transects. One transect line would be walked in an easterly direction and the personnel would continue 250 m beyond the finish before walking one kilometre either south or north to return along a different transect line in a westerly direction. This protocol was followed in order to reduce the undesirable effects caused by evasive movement of animals following disturbance during the preceding transect.
Data Analysis
The conventional distance sampling engine packaged within the specialist distance sampling software program Distance was used for analysis (Thomas et al. 2006). The process recommended by Buckland et al (2001) was followed with the transect lines defined as the sampling units. A series of plausible models combined with expansion terms were fitted to the data. A maximum of five adjustment terms were fitted using AIC by the sequential method. Histograms and qq plots were examined to assess data and model fit. In all cases, data were grouped into distance intervals, and truncation was carried out to remove outliers. The size-bias regression method was used to adjust for detection bias for clusters of animals (Buckland et al. 2001). Exact distance values, rather than distance intervals, were used in size-bias regression calculations. The non-parametric bootstrap method was used to estimate variance with resampling of 999 samples, seeded from the system clock. Confidence intervals were calculated as 2.5% and 97.5% quantiles of the bootstrap estimates. Final models were selected on the basis of the AIC, variance and chi-squared goodness of fit.
The wild mammal survey was analysed with stratification by species and habitat (vegetation class). For stratification by species, the observations for all species with a sample number of 40 or greater were examined using the detection function specific for that species. For species with an inadequate sample number for this approach, the global detection function was used. For the habitat study, the vegetation class for each observation was extracted from the classified image and density estimates made using the stratum specific detection function if the sample number was adequate. Where numbers of observations were not sufficient, data were pooled with the most similar habitat type and estimates made using the global detection function for the pooled data. Observations of rodents were not included in the analysis as they could not be identified accurately to a species level from a distance. Separate density estimates were made for warthog burrows in use at the time of the survey, as well as for the total number of warthog burrows detected (including inactive burrows).
Results
Land Cover Classification
Ten land cover classes were identified during the ground-truthing study (Table 1). The hill scrub miombo class was very small and could not be accurately mapped so was removed from the final classification (see discussion). The overall accuracy of the classification was 71.2% (95% CI 65.3–76.7%). The image produced by the fuzzy classifier was significantly more accurate than that produced using a standard maximum likelihood classifier (McNemar’s chi-squared = 4.6875, df = 1, P value = 0.030). The error matrix is presented with the conditional kappa for the classified data rather than the reference data (Table 2). The area of the park as calculated from the classified image was 331 km2 (33119 hectares) and the total perimeter length was 142 km (142102 m). The complete dataset for the final classified image (Fig. 2) is available for download via the ShareGeo open access repository at http://hdl.handle.net/10672/606.
Mammal Density
Details of observations, cluster sizes and density estimates for all species recorded during the survey are presented in Table 3. The densities of species with 40 or more observations should be considered more reliable than the other estimates as they were estimated using the stratum detection function. The overall density estimate of wild mammals in the study area, excluding rodents, was 17.32 animals/km2 (95% CI 12.69–24.59). An additional 315 rodents were observed, including 249 mopane squirrels (Paraxerus cepapi). Observations of warthogs were most frequently made in aquatic grassland and Combretum woodland with nearly a quarter of all observations made in each of these classes.
Large mammal density aggregate estimates by vegetation class are presented in Table 4. Observations for the riverine woodland class and thicket class were pooled in order to enable a class level density estimate to be made using the global detection function of the two classes. Combretum woodland and mopane scrub were similarly combined to obtain a class level estimate. Few observations were recorded for the acacia woodland and mopane scrub classes meaning that no estimate was possible for the former, and a less reliable estimate was possible for the latter, compared with other classes.
Warthog Burrow Density and Distribution
A total of 86 warthog burrows were detected during the transect survey, 42 of which appeared to have been recently used. The number of observations detected permitted an overall estimate of density, but was not sufficient to allow density estimates stratified by habitat (Table 5). The spatial distribution of warthog burrows is presented in Figure 3. The vast majority were observed in or around a slightly elevated band of Combretum woodland and thicket surrounding a large central area of mopane woodland and mopane scrub.
Discussion
Performance of the Classification
The overall accuracy of 71.2% for the final classified image was considered to be good and the fuzzy logic algorithm presented statistically significant improvements over the conventional maximum likelihood algorithm. The presence of mixed pixels in the image is likely to account for much of the difference in performance between the two. Mixed pixels have been identified as a major source of error in traditional ‘hard’ classifications that assign only one class to each pixel (Wang 1990; Foody 1996; Benz et al. 2004) and as the most important cause of misclassifications (Foody 2002). Detailed information on joint membership by other classes, particularly around boundary areas, is lost. There is no doubt the training data will have contained some mixed pixels as vegetation exists as a continuum in LNP (as in most natural ecosystems) and classes overlap. Indeed, in his detailed floristic study of North Luangwa National Park, Smith (1998) grouped his vegetation categories into mosaics of vegetation types that could not be mapped separately at his chosen resolution. Although attempts were made to collect homogenous reference data in this study, it will have contained some heterogeneity and represented, in effect, ‘fuzzy’ ground-truth data. An important feature of fuzzy classifiers is that homogenous reference data are not needed.
Most of the classes within the classification scheme performed well, with the exception of the hill scrub miombo class. A small area of this class was identified during the ground survey on the hills in the south eastern section of the park, but only at an altitude of 660 m or greater. The highest point in LNP, based on the 1:250,000 topographical maps (Surveyor-General 1972), is 680 m meaning this class was only present over a very restricted area. As it was exerting a deleterious effect on the accuracy of the rest of the classification, it was removed. Most of this area is mapped by the classification as Combretum woodland with some scrub mopane woodland. In reality, it is likely to represent more of a transition zone from Combretum woodland to scrub miombo woodland rather than just the latter.
Vegetation Composition of the Park
As discussed earlier, much of the vegetation of LNP exists in a natural mosaic of vegetation types. However, at a larger scale, several classes occur in fairly discreet zones, notably mopane woodland, mopane scrub and grassland. The two forms of Colophospermum mopane vegetation together are dominant over large areas of the park covering 37% of the total area. The large grassland habitats formed by the floodplains of the Luangwa River tributaries are a significant component of the park covering 14%. Thicket vegetation also forms fairly clear zones in places, but interdigitates with Combretum woodland in others. Aquatic grasslands, in the form of permanent or semi-permanent lagoons, account for a much smaller proportion of the total park area, but are a very characteristic feature of LNP. Riverine woodland mainly flanks the Luangwa River, but is also found in patches by the larger tributaries and lagoons. Detailed descriptions of the vegetation classes in this study may be found elsewhere (Anderson 2009).
Species Densities
Despite its small size, LNP has some distinctive wildlife habitats and supports populations of several globally threatened or endemic mammal species. The wildlife density estimates presented here represent the most detailed published information to date and can act as a baseline for on-going research and monitoring. Overall mammal densities were relatively low with some notable exceptions such as puku (Kobus vardonii).
Of the most abundant species, only warthog are known to be preferred hosts for tsetse (Clausen et al. 1998). Warthogs are generally a successful species and density estimates vary from 15 km−2 (Cumming 1975) to 30 km−2 (Estes 1993) in the best habitat (fertile alluvial soils), and less than 1 km−2 (Cumming 1975) in less favourable areas. Densities are highest in short grassland or wooded grassland areas (Rodgers 1984) and mosaics of suitable wet and dry season habitat are important (Cumming 1975). The density of warthog estimated in this study (3.14 km−2, 95% CI 1.93–5.98) was comparable with the density recorded in nearby Upper Lupande GMA (2.2 km−2) and the Zambezi Valley, Zimbabwe, but towards the lower end of reported densities (Cumming 1975; Rodgers 1984; Ndhlovu and Balakrishnan 1991). The relatively high proportion of warthog clusters observed in aquatic grassland is notable as it covered only 4% of the transect area. Aquatic grassland presents a reliable dry season source of forage for warthogs which were frequently observed digging for rhizomes of grasses and sedges in this habitat. Observations were comparatively frequent in the Combretum woodland habitat, which has a well-developed grass layer. Also notable were high densities of warthog in areas with new grass appearing after a bush fire. Marked local increases in warthog density after dry season fires have been noted before (Cumming 1975). Uncontrolled burning is a regular occurrence in LNP and is also likely to exert a significant selection pressure on the vegetation, especially given the resistant nature of Combretum species in particular to fire (Smith 1998). In turn it is likely to have significant effects on the diversity and abundance of fauna.
Warthog Burrow Distribution
Mapping of the warthog burrows over the classified dataset allowed the spatial pattern and habitat preference for burrow location to be examined. The clear pattern revealed in Figure 3 may be explained by the drainage of the soils, the ease of excavation and the provision of cover from predation. Combretum woodland and thicket are generally found on more sandy soils with better drainage in the rains and easier excavation. In contrast, mopane woodland and mopane scrub generally occur on clay soils, prone to seasonal flooding (Smith 1998) and difficult to excavate in the dry season. Warthog have been reported to thrive in areas of wooded grassland bounding suitable floodplain grassland (Rodgers 1984), a situation which occurs especially towards the south of the transect study area in LNP. The close proximity of patches of aquatic grassland to burrows makes suitable dry season grazing readily available. The close ecological association between warthog and tsetse was outlined earlier in the Introduction including the observation that apparent densities of G. m. morsitans tsetse are greatest in Combretum woodland and G. pallidipes in thicket (Anderson 2009). It is very notable, therefore, that the majority of warthog burrows are located within these two habitats.
Habitat Densities
The use of the classified dataset also allowed the aggregate density of wild mammals to be examined by vegetation class. Not surprisingly, the highest densities were recorded in grassland with nutritious herbage providing for large densities of puku in particular. Although lower densities of large mammals were recorded in the riverine woodland and aquatic grassland classes, these habitats are likely to be very important ecologically, especially in the dry season as a source of forage and water. They may support a wide diversity of other species not included in the survey such as birds, amphibians and invertebrates. Acacia woodland forms only a very small component of the vegetation in LNP and animals were rarely observed in the dense stands of Acacia kirkii, but were more commonly seen in more open acacia woodland near the Luangwa River.
It would have been desirable to estimate individual species density by habitat type, but the data were not robust enough to allow this. The large survey effort required (approximately 60 observations per habitat type for each species) makes this difficult to achieve across all habitat types, especially in environments with low mammal densities. Similarly, four land cover classes (riverine woodland, thicket, mopane scrub and Combretum woodland) did not have sufficient observations to enable the use of the stratum detection function in the analysis. Although preferable to using the global detection function, an accurate estimate was made possible by pooling the class in question with the class with the most similar visibility characteristics and using the global detection function for the two classes combined to estimate density. Riverine woodland was pooled with thicket for this purpose, and mopane scrub was pooled with Combretum woodland. Although species and cluster size may have confounded the detection probabilities to some degree, the estimated densities provide a useful indication of the general distribution of mammals.
Size of the Park
Calculation of the area of LNP using the classified image (331 km2) produces a considerably different value to the official figure for the park area (254 km2). Unfortunately, it is not clear where the official figure used by the ZAWA originates from. Changes in the course of the Luangwa River forming the western boundary of the park are occurring continuously, but will not account for such a large discrepancy. Although the exact boundary of the park is disputed by the local community, the shapefile used in this study was created through digitisation of high resolution topographical maps (Surveyor-General 1972) based on the original gazetting of the park, which suggests the national park area figures used by ZAWA may be incorrect.
Conclusion
This study provides a reliable framework for ecological monitoring of vegetation composition and wild mammal densities in remote, relatively inaccessible environments. Information generated can be used as a baseline for further study into wildlife disease systems. The use of classification algorithms based on fuzzy set theory enables accurate classification of vegetation classes despite the presence of natural mosaics and mixed pixels. The datasets created are ideal for use as a GIS base layer for the design, implementation and analysis of ecological and epidemiological studies. The distance sampling technique utilising a ground survey allows for reliable estimation of densities of smaller mammal species and important hosts of trypanosomes such as warthogs. The large survey effort required to estimate species density accurately in areas with relatively low wild mammal densities may limit the usefulness of this technique for health research in some environments.
Despite decades of research into trypanosomiasis our understanding of disease transmission in wildlife hosts is limited by the complexity and large size of the reservoir host community, and the many factors that influence it. Accurate description of the structure and distribution of communities is necessary to further our understanding and will enable better management of health relationships in remote environments such as those described in this study. Data such as these will help to enable improved modelling of disease systems with a consequential improvement in our understanding of the effects of interventions in biodiverse ecosystems.
Notes
Physiognomic refers to the overall structure or physical appearance of the community including appearance, height and spacing.
References
Anderson NE (2009) An Investigation into the ecology of trypanosomiasis in wildlife of the Luangwa Valley, Zambia Thesis. University of Edinburgh.
Anderson NE, Mubanga J, Fevre EM, Picozzi K, Eisler MC, Thomas R, et al. (2011). Characterisation of the Wildlife Reservoir Community for Human and Animal Trypanosomiasis in the Luangwa Valley, Zambia. PLoS Negl Trop Dis 5:e1211.
Astle WL (1999). A History of Wildlife Conservation and Management in the Mid-Luangwa Valley, Zambia. British Empire and Commonwealth Museum.
Astle WL, Webster R, and Lawrence CJ (1969). Land Classification for Management Planning in the Luangwa Valley of Zambia. Journal of Applied Ecology 6:143-169.
Bengis RG, Kock RA, and Fischer J (2002). Infectious animal diseases: the wildlife / livestock interface. Revue Scientifique Et Technique De L Office International Des Epizooties 21:53-65.
Benz UC, Hofmann P, Willhauck G, Lingenfelder I, and Heynen M (2004). Multi-resolution, object-oriented fuzzy analysis of remote sensing data for GIS-ready information. ISPRS Journal of Photogrammetry and Remote Sensing 58:239-258.
Buckland ST, Anderson DR, Burnham KP, Laake JL, Borchers DL, and Thomas L (2001). Introduction to distance sampling: estimating abundance of biological populations. Oxford University Press, Oxford.
Chansa W, Milanzi J, and Sichone P (2011a). Influence of river geomorphologic features on hippopotamus density distribution along the Luangwa River, Zambia. African Journal of Ecology 49:221-226.
Chansa W, Senzota R, Chabwela H, and Nyirenda V (2011b). The influence of grass biomass production on hippopotamus population density distribution along the Luangwa River in Zambia. J. Ecol. Nat. Environ 3:186-194.
Clausen PH, Adeyemi I, Bauer B, Breloeer M, Salchow F, and Staak C (1998). Host preferences of tsetse (Diptera: Glossinidae) based on bloodmeal identifications. Medical and Veterinary Entomology 12:169-180.
Claxton JR, Faye JA, and Rawlings P (1992). Trypanosome infections in warthogs (Phacochoerus aethiopicus) in the Gambia. Veterinary Parasitology, 41, 179-187, 1992.
Coates Palgrave K (2003a). Trees of Southern Africa. Struik Publishers, Cape Town.
Coates Palgrave M (2003b). Key to Some Trees of the South Luangwa Valley, Harare.
Congalton RG (1991). A Review of Assessing the Accuracy of Classifications of Remotely Sensed Data. Remote Sensing of Environment 37:35-46.
Cross P, Drewe J, Patrek V, Pearce G, Samuel M, and Delahay R (2009). Wildlife population structure and parasite transmission: implications for disease management. In: Management of Disease in Wild Mammals, Delahay R, Smith G, Hutchings M (editors), Japan: Springer, pp 9–29
Cumming DHM (1975). A field study of the ecology and behaviour of warthog. National Museum, Harare.
Dillmann JSS, and Townsend AJ (1979). Trypanosomiasis survey of wild animals in the Luangwa Valley, Zambia. Acta Tropica 36:349-356.
Estes RD (1993). The Safari Companion: A Guide to Watching African Mammals. Tutorial Press, Harare
Foody GM (1996). Approaches for the production and evaluation of fuzzy land cover classifications from remotely-sensed data. International Journal of Remote Sensing 17:1317 - 1340.
Foody GM (2002). Status of land cover classification accuracy assessment. Remote Sensing of Environment 80:185-201.
Foody GM (2004). Thematic map comparison: Evaluating the statistical significance of differences in classification accuracy. Photogrammetric Engineering and Remote Sensing 70:627-633.
Frederick H (2013). Aerial Survey Report: Luangwa Valley, 2012. Lusaka.
Kinghorn A, Yorke W, and Lloyd L (1913). Final report of the Luangwa Sleeping Sickness Commission of the BSA Co 1911-1912. Annals of Tropical Medicine and Parasitology 7:183-283.
Leak SGA (1998). Tsetse biology and ecology: their role in the epidemiology and control of trypanosomosis. In: Tsetse Biology and Ecology: Their Role in the Epidemiology and Control of Trypanosomosis. CAB International, in association with the International Livestock Research Institute, Nairobi, Kenya, Wallingford.
Lewis D, Bell SD, Fay J, Bothi KL, Gatere L, Kabila M, et al. (2011). Community Markets for Conservation (COMACO) links biodiversity conservation with sustainable improvements in livelihoods and food production. Proceedings of the National Academy of Sciences 108:13957-13962.
Lillesand TM, Kiefer RW, and Chipman JW (2004). Digital Image Processing. Pages 491-637 in Remote sensing and image interpretation. John Wiley & Sons, New York.
Marks SA (2005). Large Mammals and a Brave People, Second edition. Transaction Publishers, New Brunswick.
Michel AL, Bengis RG, Keet DF, Hofmeyr M, de Klerk LM, Cross PC, et al. (2006). Wildlife tuberculosis in South African conservation areas: Implications and challenges. Veterinary Microbiology 112:91-100.
Munang’andu HM, Siamudaala V, Munyeme M, and Nalubamba KS (2012). A review of ecological factors associated with the epidemiology of wildlife trypanosomiasis in the luangwa and zambezi valley ecosystems of zambia. Interdiscip Perspect Infect Dis 2012:372523.
NASA Landsat Programme (2004). Landsat ETM+ Scene p170r069. in.
Naylor JN, Caughley,G.J., Abel,N.O., Liberg,O. (1973). Luangwa Valley Conservation and Development Project, Zambia. FAO, Rome.
Ndhlovu DE, and Balakrishnan M (1991). Large herbivores in Upper Lupande Game Management Area, Luangwa Valley, Zambia. African Journal of Ecology 29:93-104.
Olive MM, Goodman SM, and Reynes JM (2012). The role of wild mammals in the maintenance of Rift Valley fever virus. J Wildl Dis 48:241-266.
Phiri PSM (1989) The flora of the Luangwa Valley and an analysis of its phytogeographical affinities. PhD Thesis. Reading.
Pilson RD, and Pilson BM (1967). Behaviour studies of Glossina morsitans Westwood in field. Bulletin of Entomological Research 57:227.
Plowright W, Parker J, and Pierce MA (1969). Epizootiology of African Swine Fever in Africa. Veterinary Record 85:668-674.
Pouncey R, Swanson K, and Hart K (1999). Erdas Field Guide.
Rapport DJ, Costanza R, and McMichael AJ (1998). Assessing ecosystem health. Trends in Ecology & Evolution 13:397-402.
Robinson T, Rogers D, and Williams B (1997). Univariate analysis of tsetse habitat in the common fly belt of Southern Africa using climate and remotely sensed vegetation data. Medical and Veterinary Entomology 11:223-234.
Rodgers WA (1984). Warthog Ecology in South East Tanzania. Mammalia 48:327-350.
Simukonda C (2011). Wet season survey of the African elephant and other large herbivores in selected areas of the Luangwa Valley.
Smith PP (1995). Common trees, shrubs and grasses of the Luangwa Valley. Trendrine Press St. Ives UK.
Smith PP (1998). A reconnaissance survey of the vegetation of the North Luangwa National Park, Zambia. Bothalia 28:197-211.
Somers MJ, Penzhorn BL, and Rasa OAE (1994). Home range size, range use and dispersal of warthogs in the eastern Cape, South Africa. Journal of African Zoology 108:361-373.
Stehman SV (1997). Selecting and interpreting measures of thematic classification accuracy. Remote Sensing of Environment 62:77-89.
Surveyor-General (1972) In: 1232 C1, 1232 C2, 1232 A3, 1232 A4, Lusaka.
Thomas L, Laake JL, Strindberg S, Marques FFC, Buckland ST, Borchers DL, et al. (2006). Distance 5.0. Release 2. In: U. o. S. A. Research Unit for Wildlife Population Assessment. http://www.ruwpa.st-and.ac.uk/distance/, editor.
Torr SJ (1994). Responses of tsetse flies (Diptera: Glossinidae) to warthog (Phacochoerus aethiopicus Pallas). Bulletin of Entomological Research 84:411-419.
Trapnell CG, Martin, J.D., Allan, W. (1950). A Vegetation Soil Map of Northern Rhodesia. in. Government Printer, Lusaka.
Van Wyk B, and Van Wyk P (1997). Field Guide to Trees of Southern Africa, First edition. Struik Publishers, Cape Town.
Wang F (1990). Fuzzy Supervised classification of remote-sensing images. Transactions on Geoscience and Remote Sensing 28:194-201.
White F (1983). The vegetation of Africa. In: Natural Resources Research No 20, p 50 Unesco.
Wilbroad C, and Milanzi J (2010). Population status of the hippopotamus in Zambia. African Journal of Ecology 49:130–132.
Wilkinson PJ, Pegram RG, Perry BD, Lemche J, and Schels HF (1988). The Distribution of African Swine Fever Virus Isolated From Ornithodorus-moubata In Zambia. Epidemiology and Infection 101:547-564.
Yang J, and Prince SD (2000). Remote sensing of savanna vegetation changes in Eastern Zambia 1972-1989. International Journal of Remote Sensing 21:301-322.
Acknowledgments
We gratefully acknowledge support from the Zambian Wildlife Authority and the Department of Veterinary Services, Government of Zambia. Funding was provided by the Department for International Development Animal Health Programme (DFID-AHP) and the Royal Zoological Society of Scotland. NEA and SCW are funded with support from the Ecosystem Services for Poverty Alleviation Programme (ESPA), NERC Project No. NE-J001570-1. EMF is funded by the Wellcome Trust (085308). Prof. S. Buckland provided advice regarding the design of the transect survey and Kepson Chansa assisted with the field survey. James Milansi, Wilbroad Chansa, Victor Siamudaala and Adrian Carr provided support and advice during the fieldwork.
Author information
Authors and Affiliations
Corresponding author
Additional information
Joseph Mubanga—deceased.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Anderson, N.E., Bessell, P.R., Mubanga, J. et al. Ecological Monitoring and Health Research in Luambe National Park, Zambia: Generation of Baseline Data Layers. EcoHealth 13, 511–524 (2016). https://doi.org/10.1007/s10393-016-1131-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10393-016-1131-y