Abstract
Monitoring rare and elusive species is critical in guiding appropriate conservation management measures. Mammalian carnivores are challenging to monitor directly, due to their generally nocturnal and solitary lifestyle, and relatively large home ranges. The European mink Mustela lutreola is a critically endangered, small, semi-aquatic carnivore and is one of the most threatened mammal species in Europe. In northern Spain, the European mink population is monitored regionally using different methods and approaches, making assessment of national population status difficult. There is an urgent need to 1) assess the efficacy of survey methods and 2) identify a standard monitoring methodology that can be deployed rapidly and inexpensively over large areas of the mink’s range. We deployed four methods—camera trapping, hair tubes, live trapping, and environmental DNA (eDNA) from water samples—to compare the probability of detecting European mink when present at 25 sampling sites within five 10 × 10 km2, and the economic cost and time required for each method. All four methods successfully detected European mink but the probability of detection varied by method. Camera trapping and hair tubes had the highest probability of detection; however, eDNA and live trapping detected mink in one 10 × 10 km2 where the latter two methods did not. For future European mink monitoring programs, we recommend a combination of at least two methods and suggest that camera traps or hair tubes are combined with live trapping or eDNA (depending on the scale and aims of the study), to gather critical information on distribution, occupancy and conservation status.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Monitoring rare and elusive species is critical in guiding local conservation management measures (Campbell et al. 2002; Scheele et al. 2019). This is particularly important when monitoring populations or species that are newly reintroduced into the wild, or those that are subject to change, where the reliable and rapid collection of occupancy and distribution data is vital to establishing both their short- and long-term viability. The success of these monitoring programs is, however, dependent on the monitoring method used. Key considerations are the behavior and ecology of the species (Sales et al. 2020a) and, since funding will almost always be a limiting factor, the economic cost, effort required, and likely detection success (Barea-Azcon et al. 2006, Descalzo et al. 2021).
Mammalian carnivores are a particularly challenging group to monitor directly (e.g., using non-invasive direct observations) over large spatial scales because they are generally nocturnal, solitary and have relatively large home-ranges (Barea-Azcon et al. 2006). The challenge is even greater if the species occurs at low density or is patchily distributed, whether due to natural rarity or population decline. European mink Mustela lutreola are a critically endangered, semi-aquatic carnivore, and one of the most threatened mammal species in Europe (Maran et al. 2016). European mink were historically widespread across Europe until the nineteenth century, but their total population size has declined by more than 90% since the beginning of the twentieth century (Maran et al. 2016) and now probably occupies less than 3% of their former range (Harrington et al. 2018). The current distribution of European mink is restricted to small and isolated populations in three areas: Russia; the Danube Delta in Romania and Ukraine; and northern Spain and western France (Maran et al. 2016; Zuberogoitia et al. 2018). Small populations have also been introduced to Hiiumaa Island, Estonia (Maran et al. 2017), Kunashir Island in the Russian Far East (Kisleyko et al. 2022) and Lower Saxony, Germany (Lüers and Brandt 2014). The most significant threats to European mink, wherever the species has survived, are competitive exclusion by the invasive American mink Neovison vison (Sidorovich 2001; Põdra et al. 2013), and, to a lesser extent, habitat loss and degradation and non-natural mortality such as road kills (Palazón et al. 2012, Zuberogoitia et al. 2018, reviewed in Maran et al. 2017).
In Spain, the European mink population occupied 1,900–2,000 km of watercourses in the north of the country in the early 2000s (Palazón et al. 2002). Since then, the European mink distribution area has decreased significantly due to the expansion of American mink (Põdra and Gomez 2018) and their total population size is estimated to be only a few hundred individuals (Gómez and Põdra, unpublished data). The Spanish population is probably no longer connected with the smaller French population (estimated at < 250 individuals in 2014; Direction Régionale de l’Environnement et al. 2021) and is considered perhaps only the second viable European mink population in its global range, after that in the Danube Delta (Maran et al. 2017). American mink are present in one-third of the European mink’s range in Spain, and despite control efforts since the late 1990s and early 2000s (Põdra et al. 2013; Mañas et al. 2016; Maran et al. 2017) American mink distribution continues to expand (Põdra and Gómez 2018).
Systematic monitoring of European mink is critical for assessing changes in the distribution, abundance and status of their populations, and for informing effective management strategies. Currently, in Spain, European mink are monitored by regional governments using different (direct and indirect) methods and approaches, which makes assessing the status of their population on a national scale difficult. The presence of American mink presents an extra challenge for monitoring European mink in areas where the two species co-occur as their physical similarities make distinguishing between the two species difficult (footprints detected, or photos/videos taken by camera traps in particular). European polecats Mustela putorius might also potentially be confused with European mink, although the facial markings of the two species are quite different, and polecats appear to be scarce and are rarely detected in northern Spain. To date, there has been no published scientific comparison of the relative efficacy of the different methods available to monitor European mink and no consensus on which might be the most effective and accurate. As such, there is an urgent need to assess the efficacy of survey methods and provide recommendations on a suitable standard methodology which could be applied across the mink’s Spanish range and elsewhere.
Detection rates vary among methods used to monitor wildlife populations and many methods have not been compared or validated in terms of their efficacy or reliability (Diggins et al. 2016; Witmer 2005). Currently, live trapping and camera trapping are the main methods used for monitoring European mink in Spain (e.g., Palazón et al. 2002; Gómez et al. 2011; González-Esteban et al. 2004; Põdra 2021), but the relative efficacy of these methods in reliably detecting European mink has never been directly compared. Hair tubes (sections of plastic pipe with adhesive patches to capture hair of animals that enter) have also previously been used to survey European mink and have been widely used for European pine marten Martes martes, a similar sized carnivore (Mullin et al. 2010). Environmental DNA (eDNA)-based monitoring has not yet been successfully used to detect European mink (a study in France failed to detect European mink using this approach; Steinmetz et al. 2018). However, eDNA has been successful in detecting other carnivores and semi-aquatic species (Sales et al. 2020a; Broadhurst et al. 2021). The emergence of eDNA has revolutionized biodiversity monitoring in marine and freshwater ecosystems (Deiner et al. 2017) and has been successfully applied for detecting a broad range of semi-aquatic and terrestrial mammals in both lentic and lotic systems (e.g., Harper et al. 2019; Seeber et al. 2019; Sales et al. 2020a, b; Broadhurst et al. 2021). eDNA metabarcoding has been shown to be comparable to or even outperform traditional survey methods such as field signs or camera traps for monitoring several mammal species (Fediajevaite et al. 2021; Sales et al. 2020a, b; Lyet et al. 2021; Jamwal et al. 2021) and therefore has the potential to be an effective additional method to existing monitoring approaches for European mink (Sales et al. 2020a; Broadhurst et al. 2021).
The aim of this study was to compare the efficacy of different survey methods for the critically endangered European mink with a view to identifying the most suitable candidate method for a standardized survey approach to determine the occupancy and distribution of the species. A further aim was to provide comparative data to aid in the interpretation of existing monitoring information obtained from various different methods and, in particular, to explore the potential of eDNA as an additional tool that could be used to provide range-wide distributional data. We deployed four methods—camera trapping, hair tubes, live trapping and eDNA from water samples—in northern Spain on an extant European mink population. We evaluated and compared the probability of detection and the economic cost and time required for each method. Finally, we discuss the advantages and disadvantages of each method and make recommendations for future surveys and monitoring studies.
Materials and methods
Study area
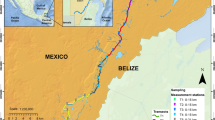
The study was carried out in north-eastern Spain in the regions of La Rioja and Álava. The European mink has been detected in only a small part of Aragon (Gómez et al. 2011) and is less abundant here than in other parts of its range. The status of European mink in La Rioja and Álava has worsened considerably in recent years, due to the invasion of American mink, but successful removal of American mink during the project LIFE LUTREOLA SPAIN (2014–2019) has enabled European mink to persist there. American mink are only detected in the area occasionally, likely comprising dispersing animals from other rival catchments. A small number of captive-born European mink (n = 15) was released in La Rioja and Álava in 2018 to reinforce the population in the Ebro basin (http://lifelutreolaspain.com, Põdra et al. unpublished data).
The surveys took place on the river Ebro, its tributaries and connected wetlands: Salburua wetland, Ea-Tiron, Oja, Najerilla, Leza, Tirón, and Zadorra (Fig. 1). The altitude is between 400 and 520 m. The Ebro river is approximately 30–100 m wide in this area, with tributaries varying between 5 and 15 m. The area of Salburu wetland is 200 ha. The region is characterized by a continental Mediterranean climate or transition from Mediterranean to Atlantic (Álava) with wetland, lakes, canals and small rivers, poplar groves and narrow riparian forests formed mostly by Salix sp, Populus sp and Alnus glutinosa, on the river banks. During the study, temperatures in the area ranged from 11 to 20 °C. A period of heavy rain occurred between the first and second eDNA water sampling sessions. During this time, the Rio Ebro was in flood conditions and water levels rose by over 1 m.
Study design
Fieldwork and data collection for all four methods were completed during October–November 2019 (post-breeding and post-dispersal for European mink) and thus overlapped temporally. Data collection for all four methods was carried out in the same five 10 km × 10 km squares based on the Universal Transverse Mercator (UTM) grid reference system. The UTM squares selected represent rivers throughout the whole Spanish European mink distribution area. Within each UTM square, five sampling sites were selected, each separated from the adjacent site by approximately 1 km of river (spacing selected to maximize the chance of having at least one site present in even the smallest female mink’s home range; previously estimated to be between 0.6 and 3.6 km (Garin et al. 2002), and 2 km (Palomares et al. 2017)). This resulted in a total of 25 sampling sites at which the methods were deployed (Fig. 1). Each sampling site was alongside a river or stream bank and sites differed in their characteristics, from main river systems to small streams to marginal backwater.
Camera trapping
Two camera traps (a Bushnell Trophy Cam HD and a Browning Strike Force HD Pro X) were deployed in a pair at each site. The rationale for deploying two cameras was to maximize the chance of capturing images with a clear view of the mink’s face (necessary for distinguishing between European and American mink) and to compare the efficacy of each camera model. The cameras were deployed either on a stake pushed into the ground, or tied to a tree, at up to 1 m above the ground (Fig. 2a). Sardines in oil were used as bait and placed on the ground or on a tree stump or log at a distance of up to 1 m from the camera. Cameras were set to record still images. Browning cameras were set to “RPF-4Shot” (to take four photos spaced 3 s apart) and the Bushnell cameras were set to take three photos per trigger. Cameras were active for 10 nights and were revisited after 5 days to replenish the bait and replace the SD cards in the cameras. All images were screened and species recorded were identified by a combination of volunteers and the authors. Species were recorded to species level, with the exception of small rodents Rodentia sp., martens Martes sp., and deer Cervidae sp., because these groups of species were not pertinent to the study. All images identified as mink were checked by the authors to confidently confirm the species (European or American).
Four monitoring methods used to detect European mink in north-eastern Spain: A Two camera traps in situ, B A hair tube, C European mink recorded by a camera trap, D European mink hair caught on the glue patch in a hair tube, E European mink being released from a live trap, F A water sample being taken for eDNA sampling
Hair tubes
Hair tubes were deployed in pairs approximately 50–100 m apart at each site, giving a total of 10 hair tubes per UTM square. Hair tubes comprised PVC tubes measuring 250 mm length and 75 mm diameter. Two adhesive patches (“mouse glue”; product code: STV182, STV International Ltd., Forge House, Little Cressington, Thetford, Norfolk, IP25 6ND, UK) were attached to the inside of the tube to collect hair from any animal that entered. Hair tubes were fixed vertically, approximately 15 cm from the ground, to either vegetation, such as shrubs or small trees adjacent to the watercourse, or to a metal or wooden rod or stake, with the open end of the tube directed towards the ground. A chicken wing was fixed inside at the top of the tube as bait.
Hair tubes were active for 10 nights and were checked daily. When a hair sample was found in the tube, it was removed, stored in ethanol and frozen. The adhesive patches were then replaced and the bait was replenished every 5 days. Guard hairs were examined under a microscope and identified to species level using characteristics as described in Teerink (1991) and González-Esteban et al. (2006). The hair of European mink can be differentiated from most mammals quite easily and this has been found to be a very reliable method (González-Esteban et al. 2006).
Live trapping
Metal cage traps were deployed in pairs approximately 50–100 m apart at each site. Traps were located within a few meters of the hair tube locations, but the trapping was carried out after the hair tube surveys were completed. Traps were baited with sardines in oil and a whole raw hen’s egg. Traps were located at least 1 m from the water and were not set during periods of heavy rain. Traps were covered with dry grass and either a wooden board or tree bark, to provide protection from the weather. The traps were set for 10 nights, although some traps were closed before the end of the 10 nights, when the water level increased, or when the same individual mink or non-target species was caught repeatedly. Traps were checked daily early in the morning.
When a European mink was captured, it was kept in the trap and covered by cloth, and transported by car to the lab (a journey of less than one hour). Mink were anaesthetized intramuscularly with a combination of 5 mg/kg ketamine hydrochloride (Imalgène 1000, Merial, Lyon, France) and 0.10 mg/kg medetomidine hydrochloride (Domtor, Orion Corporation, Espoo, Finland). While the animal was sedated, its sex was determined, morphometric measurements recorded, and a rooted hair and a 1-cm × 1-cm piece of cartilage (cut by the veterinarian) taken for genetic analysis as part of a larger study to assess the status of the European mink population in the area. A subcutaneous microchip was fitted for future identification. Sedation was reversed using Atipamezole (Antisedans, Orion Corporation, Espoo, Finland) administered intramuscularly, at five times the medetomidine dose. Mink were observed while they recovered from the anaesthetic, and released at the site of capture within two hours of processing. Other non-target animals captured were released immediately without handling.
eDNA sampling
Water samples were collected using sterile 500-ml water bottles from the bankside at a reachable distance with complete or near-complete submersion of the bottle beneath the surface. Five 500 ml water replicates (Broadhurst et al. 2021) were collected from each site (see description of camera trapping above), within 5–10 m of each other, in close proximity to the camera traps. The first eDNA sampling session was conducted between October 14th and 18th 2019 and then repeated at the same sites between October 24th and 28th 2019, covering the period when the camera traps were active. Samples were filtered on the same day as collection (along with field controls consisting of 500 ml of distilled water to test for cross-contamination during sampling) using sterilized single-use syringes and 0.45 μm Sterivex filters (Merck Millipore, Darmstadt, Germany), at a facility close to the field sites. The filters containing eDNA were transported in cool boxes with ice packs and then stored at − 20 °C until DNA extraction.
Four additional 500 ml replicates were taken from a pond within a captive European mink enclosure to test the ability of the method to detect European mink where their presence was known. These samples were transported separately from the field samples.
eDNA laboratory methods and bioinformatics
DNA was extracted from the filters in a dedicated eDNA clean room following the Mu-DNA protocol (Sellers et al. 2018). Field controls were extracted first, followed by the field eDNA samples. After the field samples were processed, the samples from the captive pond were extracted. Eight DNA extraction negative controls (one for each day of extractions) containing only extraction buffers were also included. All surfaces were sterilized with 10% bleach and then washed with 70% ethanol. Small tools were placed in a UV Stratalinker® before, in-between and after extracting each sample to reduce the risk of cross-contamination.
DNA extracts were stored at − 20 °C until PCR amplification. Eluted eDNA was amplified using the MiMammal 12S primer set (MiMammal-U-F, 5′- GGGTTGGTAAATTTCGTGCCAGC-3′; MiMammal-U-R, 5′- CATAGTGGGGTATCTAATCCCAGTTTG-3′; Ushio et al. 2017) targeting a ~ 170 bp amplicon from a variable region of the 12S rRNA mitochondrial gene with sample-specific multiplex identifier (MIDs) tags. PCR amplification protocols followed Sales et al. (2020a) with PCR positive controls (i.e., DNA extraction from a non-target species that is not locally present, the northern muriqui Brachyteles hypoxanthus from Brazil included; Broadhurst et al. 2021). Sequencing was performed over two runs using the Illumina MiSeq v2 Reagent Kits for 2 × 150 bp paired-end reads (Illumina, San Diego, CA, USA). The bioinformatic analysis was conducted using OBITools metabarcoding package (Boyer et al. 2016) following the protocol described in Sales et al. (2020a) and Broadhurst et al. (2021). See the Supplementary Material for full details of the laboratory and bioinformatic methods.
Statistical analysis
We used a Bayesian statistical analysis approach to estimate the detection probabilities of European mink for each method. We estimated the probability of detecting European mink when present throughout the study area using the R (v. 4.1.2; R Core Team 2021) package ubms (Kellner et al. 2021) and STAN software (Carpenter et al. 2017) implemented in R Studio (v. 1.2.5042; R Studio Team 2020) with 100,000 iterations, 5 chains and the default burn-in setting of half the number of iterations. The probability of detection of European mink eDNA in a sample replicate was also calculated using the R package eDNAoccupancy (Dorazio and Erickson 2018). Finally, we calculated the probability of detection for the combined use of all possible pairs of methods to assess which combination of methods offered the highest probability of detection.
McNemar’s test was used to compare the efficacy of the two camera models, using the camera model as treatment (Browning) and control (Bushnell).
Economic cost and time analysis
Costs were calculated for each method, inclusive of all aspects of fieldwork and data collection (purchasing of equipment and consumables for fieldwork, and filtering, shipping and lab costs for eDNA sampling). The time input for each method was estimated, including fieldwork, sample collection, lab work, and data analysis. Costs were calculated in Euros (€) and where costs were incurred in GDP (£), these were converted to Euros (€) in October 2021 using the rate of £1 = €1.18, to allow for consistent comparison.
Results
Camera trapping
In total, 81,655 camera trap images were recorded and at least 14 mammalian species or groups were detected; red fox Vulpes vulpes, European and American mink, Eurasian otter Lutra lutra, European pine marten, stone marten Martes foina, least weasel Mustela nivalis, common genet Genetta genetta, domestic cat Felis catus, Eurasian beaver Castor fiber, red squirrel Sciurus vulgaris, small rodent Rodentia sp. (classified to group level only), deer Cervidae sp., and wild boar Sus scrofa. Of all of the images recorded, 938 (1%) images were of mink species. Of these, 85% (n = 797) of images were confidently identified as European mink, at 48% (n = 12) of sampling sites, in three UTM squares (Fig. 3). Ten images were identified as American mink from one instance at one site. For the remaining 131 images, confident identification of the species was not possible, as the distinguishing features on the face of the mink were not fully visible.
The probability of detection of European mink per sampling site by camera trapping was 0.85, with a Bayesian Credibility Interval ranging from 0.51 to 0.99 (Fig. 4b). The average number of days to first detection was 5 (Table S1, Fig. 4a). There was heterogeneity in the number of detections of European mink at a camera trap site, ranging from 1 to 8 (mean = 2.8). The activity of European mink at the camera traps was cathemeral, with 49% of images recorded during the night and 51% recorded during daylight hours.
The Browning cameras detected European mink at 12 sites (every site where mink were detected by cameras) and the Bushnell cameras detected European mink at eight sites. Therefore, there were four sites where the Brownings detected European mink and the Bushnells did not, and no sites where the situation was reversed. Nevertheless, this effect was not statistically significant (McNemar’s, p = 0.134, χ2 = 2.25, df = 1).
a The number of days to the first detection of European mink by four monitoring methods. b The probability of detection of European mink when present for each method with their 95% Bayesian Credibility Interval. For eDNA sampling, this is represented as the average number of replicates per site (out of a total of 5 replicates per site) to the first detection
Hair tubes
A total of 31 hair samples were collected from the 25 pairs of hair tubes. Of these, 21 samples (68%) were identified as European mink; these were collected from 13 unique hair tubes at 40% (n = 10) of sampling sites, in three UTM squares (Fig. 3). The remaining samples were identified as common genet (26%), domestic cat (3%), and brown rat Rattus norvegicus (3%).
The probability of detection of European mink by hair tubes was 0.85, with a Bayesian Credibility Interval ranging from 0.51 to 0.99 (Fig. 4b). The average number of days to first detection was 3.9 (Table S1, Fig. 4a). The number of detections of European mink at a hair tube site ranged from 1 to 4 (mean = 2.1).
Live trapping
European mink were caught on 12 occasions at 40% (n = 10) of sampling sites, in four UTM squares (Fig. 3). Ten individual mink were caught; five males and five females. All individuals were caught on one occasion, with the exception of one female which was caught on three occasions in three different traps.
The probability of detection of European mink by live trapping was 0.66, with a Bayesian Credibility Interval ranging from 0.35 to 0.91 (Fig. 4b). The average number of days to first detection was 5.3 (Table S1, Fig. 4a). The number of detections of European mink at a live trapping site ranged from 1 to 2 (mean = 1.2).
eDNA sampling
The two MiSeq sequencing runs yielded a total of 19,091,872 raw sequence reads. Following the quality control and filtering steps outlined previously, 2,210,392 reads were assigned to 26 wild mammalian species and 376,580 reads (17% of total reads) were attributed to European mink. The wild mammalian species detected comprised European mink, red fox, European badger Meles meles, otter, weasel, genet, beaver, yellow-necked mouse Apodemus flavicollis, wood mouse Apodemus sylvaticus, brown rat, black rat Rattus rattus, Algerian mouse Mus spretus, edible dormouse Glis glis, Eurasian harvest mouse Micromys minutus, bank vole Myodes glareolus, common vole Microtus arvalis, field vole Microtus agrestis, Lusitanian pine vole Microtus lusitanicus, greater white-toothed shrew Crocidura russula, common shrew Sorex araneus, Aquitanian mole Talpa aquitania, European rabbit Oryctolagus cuniculus, Granada hare Lepus granatensis, red deer Cervus elaphus, roe deer Capreolus capreolus and fallow deer Dama dama. All four replicates from the captive pond were positive for European mink. The species was detected at 36% (n = 9) of sampling sites across the two sampling sessions, in four UTM squares (Fig. 3). Most mink eDNA detections occurred in the second sampling session (70% in the second sampling session; 30% in the first sampling session; Fig. 5).
The probability of detection of European mink by eDNA was 0.66, with a Bayesian Credibility Interval ranging from 0.35 to 0.91 (Fig. 4b), while the probability of detection in a replicate was 0.10 (BCI: 0.04–0.24). The average number of water sample replicates per site to first detection of mink was four (Table S1, Figs. 4b, a). All positive sites had an eDNA detection in just one sampling session, with the exception of one site at which mink were detected in both sampling sessions. Out of a total of five water sample replicates taken per site, the number of replicates that were positive for European mink eDNA ranged from 1 to 3, with the majority of sites having one positive replicate (mean = 1.3).
Combined use of two methods
We found that the combination of any two methods offered a very similar probability of detection for European mink (0.77; CI:0.46–0.97; Table S2) with the exception of the use of camera trapping and hair tubes which achieved a higher probability of detection per sampling site when present (0.85; CI:0.51–0.99); however, these two methods did not detect European mink in one UTM square where eDNA and live trapping did.
Comparison of cost and time
The most expensive method in terms of both cost and time incurred was camera trapping (Table 1). This was due to the considerable cost of purchasing 50 camera traps and the extensive time required to review and classify the camera trap images. eDNA sampling was the second most expensive in terms of cost and time, due to the costs of the lab work (extractions, PCR and sequencing) and the time required for sampling, filtering and lab analysis. Hair tubes and live trapping were the most cost-effective of the four methods (Table 1). This was due to the comparatively shorter time required and the low costs of the field equipment, which was particularly the case for hair tubes, making it the most inexpensive method overall.
Discussion
Across its global range, the population status and fine-scale distribution patterns of European mink are largely unknown. This knowledge gap is due predominantly to an absence of monitoring (e.g., across Russia and in the Danube Delta; Harrington and Maran in press) but even in areas, such as Spain, where there is considerable monitoring effort, difficulties in assessing population status and distribution arise due to inconsistent approaches among regions, a lack of consensus on which methods are most effective and accurate, and the overall difficulty in detecting European mink. We aimed to address this gap, by comparing the efficacy of different methods to inform recommendations for a standardized survey approach for European mink.
In this study, all four surveying methods successfully detected European mink at least once, but the probability of detection varied by method, with wide and overlapping Bayesian Credibility Intervals. Camera trapping detected European mink at the most sampling sites and had the joint highest probability of detection, with hair tubes. The number of unique detections by camera varied per site, with mink detected on more than one occasion at the majority of sites. The camera trap set up worked well in capturing images with a clear view of the animal’s face, allowing reliable discrimination between European or American mink; it was also the only method that detected American mink during the study. Detection of American mink is critical in any European mink monitoring strategy because understanding the distribution of American mink, and early warning of their presence, is critical to informing control efforts to protect European mink (e.g., Põdra et al. 2013). American mink are regularly caught in live traps elsewhere, and at other times in this study area, and detected by hair tubes (Põdra, pers. comm.); whether or not camera traps are truly more efficient at detecting their presence at low densities requires further study. A considerable disadvantage of camera trapping is that it is not species-specific and produced a high proportion of detections of non-target species, with images of European mink comprising only just over 1% of the total images recorded. As such, reviewing all of the images took a considerable amount of time, although this could potentially be reduced in future studies through the use of a citizen-science-based initiative or emerging machine learning algorithms to facilitate image classification (Norouzzadeh et al. 2018; Green et al. 2020; Tabak et al. 2019). As well as requiring the highest time input, camera trapping was also the most expensive method, although the cost would be reduced considerably if cameras were not purchased solely for the study and were used for repeat surveys.
This is the first study to confirm the potential of eDNA for monitoring European mink. European mink eDNA was detected at the fewest sampling sites and was typically detected in one water sample replicate out of five. As European mink are a semi-aquatic species, they would seem to be an ideal candidate for eDNA-based monitoring, although they were not detected by eDNA during a previous study in France (Steinmetz et al. 2018). However, other semi-aquatic carnivores such as the Eurasian otter have proven to be challenging to detect using eDNA, either not being detected at all in sampled areas when presence was known (Sales et al. 2020a), or requiring the screening of a comparatively large number of samples to be detected (Broadhurst et al. 2021), or requiring multiple water sample replicates to achieve a positive detection (Jamwal et al. 2021). Carnivores generally have a lower detection rate by eDNA than other mammalian species groups (Broadhurst et al. 2021; Lyet et al. 2021), with their ecologies (more solitary and wide-ranging) being proposed as the primary reason (Sales et al. 2020a). Despite these caveats, eDNA (and live trapping) detected mink in more 10 km × 10km squares than did camera trapping or hair tubes, and eDNA was successful in detecting mink at one sampling site where the species was not detected by any other method, suggesting that eDNA (and live trapping) might be more effective than other methods at a wider landscape scale.
Overall, our results add to other studies which have demonstrated the potential of eDNA metabarcoding for monitoring rare or elusive species (Franklin et al. 2019; Sales et al. 2020b), but it should be combined with other methods to maximize the chances of detecting European mink, particularly where they are at a low density (Sales et al. 2020a). The majority of European mink eDNA samples were detected in the second sampling session (Fig. 5), which may be due to heavy rain which occurred between the first and second sampling session. Lyet et al. (2021) reported a similar finding of an increased number of mammalian species detected after rainfall using eDNA and this may be due to increased river flow/transportation and run-off from the terrestrial ecosystem into the adjacent aquatic environment. The cost of eDNA sampling was relatively high and is unlikely to change for future monitoring programs in the short-term, due to the cost of field and lab materials, and the considerable staff time required for sampling, filtering and lab/bioinformatic analyses. However, a metabarcoding (multi-species) approach was used in this study which is more expensive than a species-targeted approach using either qPCR or droplet digital PCR (ddPCR). Currently, there are no available targeted assays for European mink, however a newly designed targeted eDNA approach could be developed (using the more sensitive ddPCR for example; Mauvisseau et al. 2019). Not only would this approach be less expensive, but species-targeted approaches also tend to outperform metabarcoding for detecting a focal species (Harper et al. 2019; Wood et al. 2020). However, the advantage of the multi-species metabarcoding approach is that it gives additional information on the wider mammalian community (26 species detected compared to 14 species/groups detected by camera traps) and the potential to co-detect the American mink (Broadhurst et al. 2021; although American mink were not detected by eDNA in this study), so the additional costs might be outweighed by the additional information provided (depending on the broader aims of the survey). For future monitoring programs, it will also be important to determine to what extent detections are influenced by eDNA transport and the presence of European mink upstream.
Hair tubes and live trapping detected European mink at the same proportion of sampling sites, although hair tubes had a higher probability of detection than live trapping. Both methods were slightly less effective (i.e., detected mink at fewer sites) than camera trapping, but more effective than eDNA, and both had the advantage of being relatively species-specific (unlike camera trapping and eDNA). However, although in this study, hair tubes performed well (and had the shortest time to detection of all four methods), other longer-term studies comparing hair tube and live trapping surveys carried out in parallel over a larger area in the La Rioja region of Spain than the current study, typically find that live trapping outperforms hair tubes in detecting European mink (Põdra, unpublished data). A further advantage of live trapping is the immediate identification and rapid removal of any American mink caught. This is not possible with any of the other methods, as it requires a significant amount of time (several weeks/months) to identify American or European mink either through lab analysis of eDNA or hair samples or by reviewing camera trap images, by which time it may be challenging to capture any American mink detected. Live trapping is also the only method which enables the assessment of the condition and health of individual mink (often an important goal when taking into account the critical status of the species) and for PIT tags, transmitters or radio-collars to be fitted for further monitoring. Nevertheless, live trapping requires experienced personnel, to reduce any risk of injury or incidental mortality to animals captured, and potentially the involvement of a veterinarian depending on the objective of the trapping. Although trapping is undoubtedly stressful for animals, it can be carried out safely by experienced professionals, and does provide essential information (for this endangered species) that is not possible to obtain from other means. As a non-invasive method, hair tubes are a more accessible method, easily implemented by fieldworkers with minimal training. Although not carried out in this study, hair samples can also be genotyped to identify individuals and infer population abundance (e.g., Vergara et al. 2014; O’Mahony et al. 2017), but this has not yet been validated for European mink in terms of the accuracy of abundance estimates obtained. The cost and time incurred for hair tubes and live trapping was relatively low, thus making them more affordable and feasible in future studies; however, if hair samples were to be genotyped, this would significantly increase the cost and time of this method.
The detectability of a species by different methods is influenced by behavioral traits which dictate the individual response of an animal to the method (Merrick and Koprowski 2017). Bold, active, exploratory or aggressive individuals might be more likely to explore and be detected by novel objects, such as traps and hair tubes (Carter et al. 2012), while these methods may fail to detect less active, neophobic or wary individuals (Merrick and Koprowski 2017). If this were the case for European mink, live trapping and hair tubes may fail to detect neophobic individuals, and these individuals may only be detected by eDNA sampling and camera trapping. In this instance, using a combination of methods may maximize the detection of a range of individuals and behavioral types. Our results also suggest that the relative efficacy of the survey methods tested here may differ according to the abundance of mink present, and may offer different advantages over different scales (for example, eDNA might be particularly useful for detecting mink presence over much larger areas prior to identifying sites for more detailed monitoring; Sales et al. 2020a). However, since we were only able to compare methods over a relatively small spatial scale (five discrete UTM squares), further study over a broader scale is required to verify the patterns observed here.
Recommendations and conclusion
The outlook for European mink remains perilous across its range. Accurate and repeatable information on the distribution and conservation status of extant and reintroduced populations is crucial and is needed fast before it is too late. Our study provides useful insight into some of the advantages and disadvantages of different methods, their suitability at different scales, and relative cost effectiveness. We urge researchers and conservation practitioners to work together to ensure not only that the most appropriate methods are used (dependent on the aims and scale of the survey) but also, and most importantly, that the methods used are comparable among regions and over time.
For future surveys or monitoring programs for European mink, the choice of method(s) should be dictated by the exact objectives of the study and ideally use a combination of at least two methods, in order to maximize detectability of mink. Our results suggest that both cameras and hair tubes provide effective non-invasive methods, with cameras detecting mink at the highest number of sampling sites and providing additional data on other species, although being considerably more expensive and slower to the first detection of European mink. However, to ensure detection of mink at low population densities, our results also suggest that it is important that these methods are combined with either live trapping or eDNA sampling (depending on the aim of the survey) to accurately map distribution and collect reliable occupancy data. Live trapping is a well-established method for European mink which has been used extensively in Spain, and provides individual-level data that other methods cannot that are crucial for assessing population health and viability. eDNA has potential for use over large landscape scales to determine broad distribution patterns of European mink (and thus to refine current national and global range estimates), while also providing additional presence data on a wide range of mammalian species. For eDNA sampling, we would recommend taking no fewer than five water sample replicates per site and there would likely need to be an increase in spatial and temporal resolution of samples and replicates, and the trialling of a species-specific eDNA assay (e.g., ddPCR) in comparison to multi-species (metabarcoding). For camera trapping, we would recommend using two cameras at each sampling site to maximize the chances of achieving a clear image of the mink’s face, in order to distinguish between European and American mink, and to account for possible malfunctions of the cameras. The data gathered from methods described here will be essential to inform effective, and much needed, conservation interventions for this critically endangered species.
Data availability
The datasets generated during the current study are available from the corresponding author upon reasonable request.
References
Barea-Azcón JM, Virgós E, Ballesteros-Duperón E, Moleón M, Chirosa M (2006) Surveying carnivores at large spatial scales: a comparison of four broad-applied methods. Vertebrate Conserv Biodivers 387–404
Boyer F, Mercier C, Bonin A, Le Bras Y, Taberlet P, Coissac E (2016) obitools: aunix-inspired software package for DNA metabarcoding. Mol Ecol Resour 16(1):176–182
Broadhurst HA, Gregory LM, Bleakley EK, Perkins JC, Lavin JV, Bolton P, Browett SS, Howe CV, Singleton N, Tansley D, Sales NG, McDevitt AD (2021) Mapping differences in mammalian distributions and diversity using environmental DNA from rivers. Sci Total Environ 801:149724
Campbell SP, Clark JA, Crampton LH, Guerry AD, Hatch LT, Hosseini PR, Lawler JJ, O’Connor RJ (2002) An assessment of monitoring efforts in endangered species recovery plans. Ecol Appl 12(3):674–681
Carpenter B, Gelman A, Hoffman MD, Lee D, Goodrich B, Betancourt M, Brubaker M, Guo J, Li P, Riddell A (2017) Stan: A probabilistic programming language. J Stat Softw 76:1–32
Carter AC, Heinsohn R, Goldizen AW, Biro PA (2012) Boldness, trappability and sampling bias in wild lizards. Anim Behav 83(4):1051–1058
Deiner K, Bik HM, Mächler E, Seymour M, Lacoursière-Roussel A, Altermatt F, Creer S, Bista I, Lodge DM, de Vere N (2017) Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol Ecol 26(21):5872–5895
Descalzo E, Jiménez J, Delibes-Mateos M, Díaz-Ruiz F, Ferreras P (2021) Assessment of methods for detecting an opportunistic and expanding mesocarnivore in southwestern Europe. J Zool. https://doi.org/10.1111/jzo.12912
de l’Environnement DR (2021) de l’Aménagement et du Logement (DREAL), Groupe de Recherche et d’Investigation sur la Faune Sauvage (GRIFS), Cistude Nature, Office Français de la Biodiversité (OFB). Plan National d’Actions en faveur du Vison d’Europe (Mustela lutreola) 2021–2031 174p. Available from: https://www.nouvelle-aquitaine.developpement-durable.gouv.fr/IMG/pdf/2022_01_13_3rd_nap_for_european_mink_2021-2031_.pdf
Diggins CA, Gilley M, Kelly CA, Ford M (2016) Comparison of survey techniques on detection of northern flying squirrels. Wildl Soc Bull 40(4):654–662
Dorazio RM, Erickson RA (2018) ednaoccupancy: An r package for multiscale occupancy modelling of environmental DNA data. Mol Ecol Resour 18:368–380
Fediajevaite J, Priestley V, Arnold R, Savolainen V (2021) Meta-analysis shows that environmental DNA outperforms traditional surveys, but warrants better reporting standards. Ecol Evol. https://doi.org/10.1002/ece3.7382
Franklin TW, McKelveya KS, Goldinga JD, Masona DH, Dysthea JC, Pilgrima KL et al (2019) Using environmental DNA methods to improve winter surveys for rare carnivores: DNA from snow and improved noninvasive techniques. Biol Cons 229:50–58
Garin I, Zuberogoitia I, Zabala J, Aihartza J, Clevenger AP, Rallo A (2002) Home ranges of European mink Mustela lutreola in southwestern Europe. Acta Theriol 47:55–62. https://doi.org/10.1007/BF03193566
Gómez A, Oreca S, Põdra M, Sanz B, Palazón S (2011) Expansión del visón europeo Mustela lutreola (Linnaeus, 1761) hacia el este de su área de distribución en España: primeros datos en Aragón. Galemys 23:37–45
González-Esteban J, Villate I, Irizar I (2004) Assessing camera traps for surveying the European mink, Mustela lutreola (Linnaeus, 1761), distribution. Eur J Wildl Res 50:33–36
González-Esteban J, Villate I, Irizar I (2006) Differentiating hair samples of the European mink (Mustela lutreola), the American mink (Mustela vison) and the European polecat (Mustela putorius) using light microscopy. J Zool 270:458–461
Green SE, Rees JP, Stephens PA, Hill RA, Giordano AJ (2020) Innovations in Camera Trapping Technology and Approaches: The Integration of Citizen Science and Artificial Intelligence. Animals 10(1):132. https://doi.org/10.3390/ani10010132
Harper LR, Handley LL, Carpenter AI, Ghazali M, Di Muri C, Macgregor CJ et al (2019) Environmental DNA (eDNA) metabarcoding of pond water as a tool to survey conservation and management priority mammals. Biol Conserv 238:108225. https://doi.org/10.1016/j.biocon.2019.108225
Harrington LA, Maran T (in press) European Mink. In: Handbook of the Mammals of Europe. Edited by K. Hackländer & F.E. Zachos: Springer (2024)
Harrington LA, Pdra M, Gómez A, Maran T (2018) Raising awareness of the plight of the critically endangered European mink in Spain is not miscommunication – a response to Melero. Letter to the Editor. Biodivers Conserv 27:269–271. https://doi.org/10.1007/s10531-017-1419-4
Jamwal PS, Bruno A, Galimberti A, Magnani D, Krupa H, Casiraghi M, Loy A (2021) First assessment of eDNA-based detection approach to monitor the presence of Eurasian otter in Southern Italy. Hystrix Italian J Mammal 32(2):176–181
Kellner KF, Fowler NL, Petroelje TR, Kautz TM, Beyer DE Jr, Belant JL (2021) ubms: An R package for fitting hierarchical occupancy and N-mixture abundance models in a Bayesian framework. Methods Ecol Evol. https://doi.org/10.1111/2041-210X.13777
Kisleyko AA, Dinets V, Grishchenko MY, Kozlovskiy EE, Khlyap LA (2022) The European Mink (Mustela lutreola) on Kunashir Island: Confirmed Survival 40 years After Introduction. Mammal Study 47(3):1–10. https://doi.org/10.3106/ms2021-0044
Lüers E, Brandt T (2014) Ein Versuch zur Wiedaransiedlung des Europäischen Nerzes (Mustela lutreola) am Steinhuder Meer, Niedarsachsen. Säugetierkundliche Informationen 9:249–264
Lyet A, Pellissier L, Valentini A, Dejean T, Hehmeyer A, Naidoo R (2021) eDNA sampled from stream networks correlates with camera trap detection rates of terrestrial mammals. Sci Rep 11:11362
Mañas S, Gómez A, Palazón S, Põdra M, Binobis B, Alarcia OE, Casal J, Ruiz-Olmo J (2016) Are we able to affect the population structure of an invasive species through culling? A case study of the attempts to control the American mink population in the Northern Iberian Peninsula. Mammal Res 61(4):309–317
Maran T, Põdra M, Harrington LA, Macdonald DW (2017) European mink: restoration attempts for a species on the brink of extinction. In: Biology and Conservation of Musteloids. Edited by David W. Macdonald, Chris Newman, and Lauren A. Harrington: Oxford University Press (2017). Oxford Univ Press. https://doi.org/10.1093/oso/9780198759805.001.0017
Maran T, Skumatov D, Gómez A, Põdra M, Abramov AV, Dinets V (2016) Mustela lutreola. The IUCN Red List of Threatened Species 2016:e.T14018A45199861. https://doi.org/10.2305/IUCN.UK.2016-1.RLTS.T14018A45199861.en. Accessed 31 Oct 2019
Mauvisseau Q, Davy-Bowker J, Bulling M, Brys R, Neyrinck S, Troth C, Sweet M (2019) Combining ddPCR and environmental DNA to improve detection capabilities of a critically endangered freshwater invertebrate. Sci Rep 9:14064. https://doi.org/10.1038/s41598-019-50571-9
Merrick J, Koprowski JL (2017) Should we consider individual behavior differences in applied wildlife conservation studies? Biol Cons 209:34–44
Mullin J, Statham MJ, Roche T, Turner PD, O’Reilly C (2010) Remotely plucked hair genotyping: a reliable and non-invasive method for censusing pine marten (Martes martes, L. 1758) populations. Eur J Wildl Res 56:443–453
Norouzzadeh MS, Nguyen A, Kosmala M, Swanson A, Palmer MS, Packer C, Clune J (2018) Automatically identifying, counting, and describing wild animals in camera-trap images with deep learning. PNAS 155:(25). https://doi.org/10.1073/pnas.1719367115
O’Mahony DT, Powell C, Power J, Hanniffy R, Marnell F, Turner P, O’Reilly C (2017) Non-invasively determined multi-site variation in pine marten Martes martes density, a recovering carnivore in Europe. Eur J Wildl Res. https://doi.org/10.1007/s10344-017-1108-3
Palazón S, Ceña JC, Ruiz-Olmo J, Ceña A, Gosálbez J, Gómez-Gayubo A (2002) Trends in distribution of the European mink (Mustela lutreola L, 1761) in Spain: 1950–1999. Mammalia 67(4):473–484
Palazón S, Melero Y, Gómez A, de Luzuriaga JL, Põdra M, Gosàlbez J (2012) Causes and patterns of human-induced mortality in the Critically Endangered European mink Mustela lutreola in Spain. Oryx 46(4):614–616. https://doi.org/10.1017/S0030605312000920
Palomares F, Lópex-Bao JV, Telletxea G, Ceña JC, Fournier P, Giralda G, Urra F (2017) Activity and home range in a recently widespread European mink population in Western Europe. Eur J Wildl Res 63:78
Põdra M (2021) Expansion of alien American mink, Neovison vison, and translocation of captive-bred European mink, Mustela lutreola: assessing impact on the native species’ conservation. Ph.D. Dissertation, Tallinn University
Põdra M, Gómez A (2018) Rapid expansion of the American mink poses a serious threat to the European mink in Spain. Mammalia. https://doi.org/10.1515/mammalia-2017-0013
Põdra M, Gómez A, Palazón S (2013) Do American mink kill European mink? Cautionary message for future recovery efforts. Eur J Wildl Res 59:431–440
R Core Team (2021) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
R Studio Team (2020) RStudio: Integrated Development for R. RStudio, Inc., Boston, MA. http://www.rstudio.com/
Sales NG, Kaizer MC, Coscia I, Perkins JC, Highlands A, Boubli JP, Magnusson WE, Da Silva MNF, Benvenuto C, McDevitt AD (2020b) Assessing the potential of environmental DNA metabarcoding for monitoring Neotropical mammals: a case study in the Amazon and Atlantic Forest, Brazil. Mammal Rev 50:221–225
Sales NG, McKenzie MB, Drake J, Harper LR, Browett SS, Coscia I et al (2020a) Fishing for mammals: Landscape-level monitoring of terrestrial and semi-aquatic communities using eDNA from riverine systems. J Appl Ecol 57:707–716
Scheele BC, Legge S, Blanchard W, Garnett S, Geyle H, Gillespie G, Harrison P, Lindenmayer D, Lintermans M, Robinson N, Woinarski J (2019) Continental-scale assessment reveals inadequate monitoring for threatened vertebrates in a megadiverse country. Biol Cons 235:273–278
Sellers GS, Di Muri C, Gómez A, Hänfling B (2018) Mu-DNA: a modular universal DNA extraction method adaptable for a wide range of sample types. Metabarcoding Metagen 2:e24556
Seeber PA, McEwen GC, Löber U, Förster DW, East ML, Melzheimer J, Greenwood AD (2019) Terrestrial mammal surveillance using hybridization capture of environmental DNA from African waterholes. Mol Ecol Resour 19(6):1486–1496
Sidorovich VE (2001) Study on the decline in the European mink Mustela lutreola population in connection with the American mink M. vison expansion in Belarus: story of the study, review of the results and research priorities. Säugetierkundliche Informationen 5:133–153
Steinmetz J, Ruette S, Ruys T, Jean P, Dejean T (2018) Vers une nouvelle méthode de détection des espèces de mammifères semi-aquatiques : étude pilote et approche « Metabarcoding ADNe ». Faune Sauvage 7. Available from: https://professionnels.ofb.fr/sites/default/files/pdf/RevueFS/FauneSauvage319_2018_Art2.pdf
Tabak MA, Norouzzadeh MS, Wolfson DW, Sweeney SJ, Vercauteren KC, Snow NP et al (2019) Machine learning to classify animal species in camera trap images: Applications in ecology. Methods Ecol Evol 10(4):585–590
Teerink BJ (1991) Hair of West-European mammals: atlas and identification key. Cambridge University Press, Cambridge
Ushio M, Fukuda H, Inoue T, Makoto K, Kishida O, Sato K, Murata K, Nikaido M, Sado T, Sato Y, Takeshita M, Iwasaki W, Yamanaka H, Kondoh M, Miya M (2017) Environmental DNA enables detection of terrestrial mammals from forest pond water. Mol Ecol Resour 17(6):e63–e75
Vergara M, Ruiz-González A, Luzuriaga JL, Gómez-Moliner BJ (2014) Individual identification and distribution assessment of otters (Lutra lutra) through non-invasive genetic sampling: Recovery of an endangered species in the Basque Country (Northern Spain). Mamm Biol 79:259–267
Witmer GW (2005) Wildlife population monitoring: some practical considerations. Wildl Res 32:259–263
Wood ZT, Erdman BF, York G, Trial JG, Kinnison MT (2020) Experimental assessment of optimal lotic eDNA sampling and assay multiplexing for a critically endangered fish. Environmental DNA. https://doi.org/10.1002/edn3.64
Zuberogoitia I, Põdra M, Palazón S, Gómez A, Zabala N, Zabala J (2018) Facing Extinction, Last Call for the European Mink. Ann Rev Res 2:2
Acknowledgements
The live trapping and hair tube surveys were supported by La Rioja Regional Government, the Ministry for the Ecological Transition and the Demographic challenge and Tragsatec. The eDNA component of this work was funded by grants from the Vincent Wildlife Trust, People’s Trust for Endangered Species and a University of Salford Internal Research Award given to ADM. We thank the members of the Molecular Ecology Group in Salford for advice on laboratory work and bioinformatics. We are grateful to all of the volunteers who assisted with classification of camera trap images.
Author information
Authors and Affiliations
Contributions
SC, LH, MP, ADM, JM, EC, and RH conceived and designed the study; RH, AH, MP, AG, JPR, DLA, IG, JU, and AT conducted fieldwork and data collection; PLB, JVL, and SSB performed the eDNA laboratory work and PLB and ADM analysed the eDNA data; RH, EC, JM, and PW performed data collation and statistical analyses; EC drafted the manuscript, with input from ADM and LH; all authors reviewed and approved the final version.
Corresponding author
Ethics declarations
Ethical approval
This study involved live trapping European mink, and handling of the animals under anaesthesia—these procedures were carried out as part of a larger on-going TRAGSATEC study of European mink in Spain and were carried out under license from regional governments.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Croose, E., Hanniffy, R., Harrington, A. et al. Mink on the brink: comparing survey methods for detecting a critically endangered carnivore, the European mink Mustela lutreola. Eur J Wildl Res 69, 34 (2023). https://doi.org/10.1007/s10344-023-01657-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10344-023-01657-3