Abstract
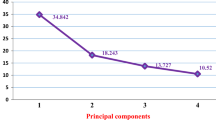
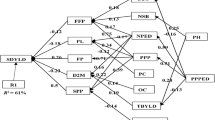
Among the edible legumes, the common bean (Phaseolus vulgaris L.) has first place in the world in terms of production quantity and plays a great role in human nutrition. There is an ever-increasing interest in organically-grown foodstuffs, especially by dieticians and conscious consumers who wish to consume chemical residue-free products. This study was conducted in the 2018 and 2019 growing seasons under organic farming conditions without any chemical inputs to determine agro-morphological traits of 16 registered common bean cultivars and 4 local genotypes. Experiments were conducted in randomized blocks design with 3 replications. For agro-morphological traits, plant height (PH), first pod height (FPH), number of branches per plant (NBP), number of pods per plant (NPP), number of seeds per pod (NSPO), number of seeds per plant (NSPL), seed yield (SY), 100-seed weight (100-SW) and seed protein content (PC) were investigated. Experimental data were assessed through multi-variate analysis methods. Correlation analysis revealed the greatest positive correlation between NPP—NSPL (r = 0.8851) and the greatest negative correlation between NSPL—100 SW (r = −0.6118). In principal component analysis (PCA) the first principal component (PC1) axis had NSPL, NPP, NSPO, FPH and 100-SW parameters and explained 41.28% of the total variation. The first 3 principal components explained 76.29% of the total variation. Hierarchical cluster analysis, performed to determine genetic relationships among the genotypes, revealed 5 main clusters and Güzelöz genotype alone formed the 5th cluster. The greatest genetic distance of cultivars was encountered between Adabeyazı—Aras cultivars (6.963) and the greatest genetic distance of local genotypes was seen between Güzelöz—Aras genotypes (4.944).
Zusammenfassung
Unter den essbaren Hülsenfrüchten, die Gemeine Bohne (Phaseolus vulgaris L.) hat weltweit den ersten Platz in Bezug auf die Produktionsmenge und spielt eine große Rolle in der menschlichen Ernährung. Das Interesse an Lebensmitteln aus biologischem Anbau wächst ständig, insbesondere von Ernährungswissenschaftlern und bewussten Verbrauchern, die Produkte ohne chemische Rückstände konsumieren möchten. Diese Studie wurde in den Vegetationsperioden 2018 und 2019 unter Bedingungen des ökologischen Landbaus ohne chemische Zusätze durchgeführt, um agromorphologische Merkmale von 16 registrierten gemeinen Bohnensorten und 4 lokalen Genotypen zu bestimmen. Die Experimente wurden in einem randomisierten Blockdesign mit 3 Wiederholungen durchgeführt. Für agromorphologische Merkmale, Pflanzenhöhe (PH), die erste Podhöhe (FPH), Anzahl der Blätter pro Pflanze (NBP), Anzahl Schoten pro Pflanze (NPP), Anzahl Samen pro Schote (NSPO), Anzahl der Samen pro Pflanze (NSPL), Samenertrag (SY), 100-Samen-Gewicht (100-SW) und Samenproteingehalt (PC) wurden untersucht. Versuchsdaten wurden durch multivariate Analysemethoden bewertet. Die Korrelationsanalyse ergab die größte positive Korrelation zwischen NPP – NSPL (r=0,8851) und die größte negative Korrelation zwischen NSPL–100 SW (r = −0,6118). In der Hauptkomponentenanalyse (PCA) hatte die erste Hauptkomponentenachse (PC1) NSPL-, NPP-, NSPO-, FPH- und 100-SW-Parameter und erklärte 41,28 % der Gesamtvariation. Die ersten drei Hauptkomponenten erklärten 76,29 % der Gesamtvariation. Eine hierarchische Clusteranalyse, die durchgeführt wurde, um genetische Beziehungen zwischen den Genotypen zu bestimmen, ergab fünf Hauptcluster, und der Güzelöz Genotyp allein bildete den fünften Cluster. Die größte genetische Distanz von Genotypen wurde zwischen Adabeyazı-Aras Genotypen (6.963) und die größte genetische Distanz lokaler Genotypen wurde zwischen Güzelöz-Aras Genotypen (4.944) festgestellt.


Similar content being viewed by others
References
Ashgari A, Vojdani P (1997) Study of genetic diversity of the Iranian common bean land races in relation to geographical. Hort Abst 1(1):549
Babagil G (2011) Erzurum ekolojik koşulları da bazı nohut (Cicer arietinum L.) çeşitleri verim ve verim özelliklerinin incelenmesi. Anadolu Tarım Bilim Derg 26(2):122–127
Bazzano LA, Thompson AM, Tees MT, Nguyen CH, Winham DM (2011) Non-soy legume consumption lowers cholesterol levels: a meta-analysis of randomized controlled trials. Nutr Metab Cardiovasc Dis 21(2):94–103
Benlioğlu B, Ozkan U (2022) Multivariate analysis and its application for screening mungbean (Vigna radiata (L.) Wilczek) landraces. Legum Res. https://doi.org/10.18805/LRF-661
Blair MW (2013) Mineral biofortification strategies for food staples: the example of common bean. J Agric Food Chem 61(35):8287–8294
Blair MW, Astudillo C, Grusak MA, Graham R, Beebe SE (2009) Inheritance of seed iron and zinc concentrations in common bean (Phaseolus vulgaris L.). Mol Breed 23(2):197–207
Blair MW, Soler A, Cortés AJ (2012) Diversification and population structure in common beans (Phaseolus vulgaris L.). Plos One 7:e49488. https://doi.org/10.1371/journal.pone.0049488
Boghara MC, Dhaduk HL, Kumar S, Parekh MJ, Patel NJ, Sharma R (2016) Genetic divergence, path analysis and molecular diversity analysis in cluster bean (Cyamopsis tetragonoloba L. Taub.). Ind Crops Prod 89:468–477
Bozoğlu H, Gülümser A (2000) Kuru fasulyede (Phaseolus vulgaris L.) bazı tarımsal özelliklerin genotip çevre interaksiyonları ve stabilitelerinin belirlenmesi üzerine bir araştırma. Turk J Agric For 24:211–220
Canci H, Bozkurt M, Kantar F, Yeken MZ, Ozer G, Cifci V (2019) Investigation and characterization of phaseolus bean bio-diversity in western Anatolia. KSU J Agric Nat 22(Suppl. 2):251–263. https://doi.org/10.18016/ksutarimdoga.vi.579482
Çalışkan S, Aytekin Rİ, Yağız AK, Yavuz C (2018) Effect of full and limited irrigation treatments on seed quality of some common bean cultivars (Phaseolus vulgaris L.). Turk J Agric Sci Technol 6(12):1853–1859
Celmeli T, Sari H, Canci H, Sari D, Adak A, Eker T, Toker T (2018) The nutritional content of common bean (Phaseolus vulgaris L.) landraces in comparison to modern varieties. Agronomy 8(9):1–9. https://doi.org/10.3390/agronomy8090166
Ceyhan E, Önder M, Karaman A (2009) Fasulye genotiplerinin bazı tarımsal özelliklerinin belirlenmesi. Selcuk J Agric Food Sci 23(49):67–73
Chen C, Chen S, Jha RK, Cotrozzi L, Nali C, Lorenzini G, Ma L (2020) Phenol metabolism of two cultivars of durum wheat (Triticum durum Desf.) as affected by ozone and flooding stress. J Agron Crop Sci 206(3):338–351. https://doi.org/10.1111/jac.12394
Elkoca E, Kantar F (2004) Erzurum Ekolojik Koşullarına Uygun Erkenci ve Yüksek Verimli Kuru Fasulye (Phaseolus vulgaris L.) Genotiplerinin belirlenmesi/determination of early maturing and high yielding dry bean (Phaseolus vulgaris L.) genotypes suitable for Erzurum ecological co. Atatürk Üniv Ziraat Fakü Derg 35:3–4
Freytag GF, Debouck DG (2002) Review of taxonomy, distribution, and ecology of the genus Phaseolus (Leguminosae papilionoideae) in North America, Mexico, and Central America. Sida Bot Misc 23:1–300
Ha V, Sievenpiper JL, de Souza RJ, Jayalath VH, Mirrahimi A, Agarwal A, Chiavaroli L, Mejia SB, Sacks FM, Di Buono M, Bernstein AM, Leiter AL, Kris-Etherton PM, Vuksan V, Bazinet RP, Josse RG, Beyene J, Kendall CWC, Jenkins DJA (2014) Effect of dietary pulse ıntake on established therapeutic lipid targets for cardiovascuar risk reduction: a systematic review and meta-analysis of randomized controlled trials. CMAJ (Ottawa) 186(8):252–262
Iwata-Otsubo A, Radke B, Findley S, Abernathy B, Vallejos CE, Jackson SA (2016) Fluorescence in situ hybridization (FISH)-based karyotyping reveals rapid evolution of centromeric and subtelomeric repeats in common bean (Phaseolus vulgaris) and relatives. Genes Genomes Genet 6(4):1013–1022. https://doi.org/10.1534/g3.115.024984
Karaagac O, Balkaya A (2013) Interspecific hybridization and hybrid seed yield of winter squash (Cucurbita maxima Duch) and pumpkin (Cucurbita moschata Duch) lines for rootstock breeding. Sci Hortic 149:9–12
Loko LEY, Orobiyi A, Adjatin A, Akpo J, Toffa J, Djedatin G, Dansi A (2018) Morphological characterization of common bean (Phaseolus vulgaris L.) landraces of Central region of Benin Republic. J Plant Breed Crop Sci 10(11):304–318. https://doi.org/10.5897/JPBCS2018.0766
Marzooghian A, Moghaddam M, Valizadeh M, Kooshki MH (2012) Genetic diversity of common bean genotypes as revealed by seed storage proteins and some agronomic traits. Plant Breed Seed Sci 67:125–137. https://doi.org/10.2478/v10129-011-0075-1
Nadeem MA, Habyarimana E, Çiftçi V, Nawaz MA, Karaköy T, Comertpay G, Shahid MQ, Hatipoğlu R, Yeken MZ, Ali F, Ercişli S, Chung G, Baloch FS (2018) Characterization of genetic diversity in Turkish common bean gene pool using phenotypic and whole-genome DArTseqgenerated silicoDArT marker information. PLoS ONE 13(10):e205363. https://doi.org/10.1371/journal.pone.0205363
Ozaktan H (2021) Technological characteristics of chickpea (Cicer arietinum L.) cultivars grown under natural conditions. Turk J Field Crop 26(2):235–243
Pathak R, Singh SK, Singh M, Henry A (2010) Molecular assessment of genetic diversity in cluster bean (Cyamopsis tetragonoloba) genotypes. J Genet 89:243–246
Pathak R, Singh SK, Singh M (2011) Assessment of genetic diversity in cluster bean using nuclear rDNA and RAPD markers. J Food Legum 24:180–183
Petry N, Boy E, Wirth J, Hurrell R (2015) The potential of the common bean (Phaseolus vulgaris) as a vehicle for iron biofortification. Nutrients 7(2):1144–1173
Singh RJ (2007) Landmark research in oilseed crops. In: Singh RJ, Jauhar PP (eds) Oilseed crops. Genetic resources, chromosome engineering, and crop improvement series, vol 4. CRC Press, Boca Raton, pp 1–12
Sinkovic L, Pipan B, Sinkovic E, Meglic V (2019) Morphological seed characterization of common (Phaseolus vulgaris L.) and runner (Phaseolus coccineus L.) bean germplasm: a Slovenian gene bank example. Biomed Res Int. https://doi.org/10.1155/2019/6376948
Smykal P, Coyne CJ, Ambrose MJ, Maxted N, Schaefer H, Blair MW, Berger J, Greene SL, Nelson MN, Besharat N et al (2015) Legume crops phylogeny and genetic diversity for science and breeding. Crit Rev Plant Sci 34(1–3):43–104. https://doi.org/10.1080/07352689.2014.897904
Steel RGD, Torrie JH (1980) Principles and procedures of statistics. McGraw-Hill, N.Y.
Tor-Roca A, Garcia-Aloy M, Mattivi F, Llorach R, Andres-Lacueva C, Urpi-Sarda M (2020) Phytochemicals in legumes: a qualitative reviewed analysis. J Agric Food Chem 68:13486–13496
Venora G, Blangifortil S, Cremonini R (1999) Karyotype analysis of twelve species belonging to genus Vigna. Cytologia 64:117–127. https://doi.org/10.1508/ytologia.64.117
Wani IA, Sogi DS, Wani AA, Gill BS (2017) Physical and cooking characteristics of some Indian kidney bean (Phaseolus vulgaris L.) cultivars. J Saudi Soc Agric Sci 16(1):7–15
Yeken MZ, Nadeem MA, Karaköy T, Baloch FS, Çiftçi V (2019) Determination of Turkish common bean germplasm for morpho-agronomic and mineral variations for breeding perspectives in Turkey. Kahramanmaraş Sütçü İmam Üniv Tarım Ve Doğa Derg 22:38–50
Funding
This study was supported by Turkish Scientific Research Council (TUBİTAK) with the project number of 119O226.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
H. Ozaktan, S. Uzun, O. Uzun and C. Yasar Ciftci declare that they have no competing interests.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ozaktan, H., Uzun, S., Uzun, O. et al. Assessment of Agro-Morphological Traits of Common Bean Genotypes Grown Under Organic Farming Conditions with Multi-Variate Analyses and Applications. Gesunde Pflanzen 75, 515–523 (2023). https://doi.org/10.1007/s10343-022-00713-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10343-022-00713-3