Abstract
Populations of smaller effective sizes or subjected to strong environmental filtering have a higher risk of stochastic changes in their genetic properties. This study tested the hypothesis that reducing the density of parent trees combined with environmental filtering during the stage of regeneration establishment affects gene flow between the parent and offspring populations of Abies alba Mill. The genetic variation and spatial genetic structure of parent trees and seedlings were compared in the Western Carpathians (southern Poland) in 27 stands with varying adult tree and regeneration densities. The analysis comprised 1592 individuals and was based on five nuclear microsatellite markers. The proportion of stands with significant spatial genetic structure in the adult tree and seedling cohorts increased as the density of adult trees decreased. The genetic differentiation between adult trees and seedlings expressed by the fixation index FST was between 0.005 and 0.021 and increased as the density of adult trees decreased but was not correlated with the seedling abundance. At the within-stand level, the analysis revealed shifts in the average number of alleles per locus (between − 15% and + 28%), the observed heterozygosity (between − 25% and + 20%) and expected heterozygosity (between − 8% and + 11%) between the populations of adult trees and seedlings. It was concluded that in mixed stands with densities of A. alba trees about 50 adult individuals per ha, there may be some risk of reduced effective population size and disturbed vertical gene flow, but significant shifts in the level of genetic variation are less likely.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Natural regeneration is commonly considered the best strategy for maintaining genetic diversity and evolutionary potential in most forest tree species with large populations (Finkeldey and Ziehe 2004; Geburek and Müller 2005). It remains questionable, however, if this strategy is equally appropriate for stands with a low density of reproducing individuals, or where micro-environmental fragmentation or strong bottleneck effects are expected to occur (Geburek and Müller 2005; Hosius et al. 2006).
A low density of parent trees may result from spatial fragmentation at the landscape level (De-Lucas et al. 2009; Leonardi et al. 2012), assortative mating for tree size or phenology (Gérard et al. 2006), or reproduction during a few flowering and seed production seasons (Sork et al. 2002). Populations of smaller effective sizes have a higher risk of stochastic changes in their genetic properties which could result from a number of factors, including a bottleneck in the census size or a large variance in reproductive output (Wang 2005). Reproductive isolation causes shifts in the mating system from outcrossing to selfing (Restoux et al. 2008), reduces the diversity of the local pollen pool and increases the relatedness in the next generation leading to mating between closely related individuals and inbreeding in subsequent generations (El-Kassaby and Jaquish 1996; Robledo-Arnuncio et al. 2004a). Reduced pollen diversity may also impose fitness costs on future generations by increasing the number of recessive deleterious alleles within the paternal mating pool and decreasing pollen competition (Breed et al. 2012). Within naturally regenerating tree populations, restricted pollen and seed dispersal can induce local genetic differentiation and the formation of family structures (Vornam et al. 2004; Rajendra et al. 2014). Spatial genetic structure (SGS) is likely to be stronger in forest stands having a low density of reproductive trees (Hamrick et al. 1993; Kyndt et al. 2009), although its formation is influenced by various factors, including gene dispersal parameters, mating patterns, and the spatial distribution of individual parents (De-Lucas et al. 2009; Sagnard et al. 2011; Hoban et al. 2014).
Gene transmission between the parent generation and its progeny is also strongly influenced by natural selection and environmental filtering. The reproductive success of adult individuals depends greatly on the coincidence of their seed crop years with the occurrence of disturbing events reducing canopy density (Beaudet et al. 2007; Bladé and Vallejo 2008), environmental heterogeneity (Snyder and Chesson 2004; Buck and Clair 2014), and the dynamics of biotic factors affecting progeny survival (Nopp-Mayr et al. 2012; Paluch and Stępniewska 2012). These factors could, by narrowing the recruitment windows or generating bottleneck effects, be a source of an imbalance in gene representation (Konnert and Hussendörfer 2001; Vranckx et al. 2014). Over a longer period, this initial imbalance may be reinforced by the increased competitive ability of the oldest regeneration cohorts (McClure et al. 2000; Madsen and Hahn 2008). Nevertheless, these processes are highly stochastic, and their cumulative influence on genetic diversity and SGS are usually difficult to predict (De Andrés et al. 2014).
This study compared the genetic structure of adult trees and their progeny in naturally regenerating Abies alba Mill. populations in the Western Carpathians (southern Poland). A. alba is a late successional conifer tree species, which frequently appears in stands consisting of Fagus sylvatica L. and/or Picea abies (L.) H. Karst. In silvicultural practice, this species is mostly regenerated naturally using shelterwood, irregular shelterwood, or single-tree selection systems. However, in spite of its high shade tolerance and growth plasticity, its regeneration tends to accumulate slowly under the parent stand canopy, often forming irregular spatial distribution patterns that indicate strong habitat fragmentation and environmental filtering (Paluch 2005). In the Western Carpathians, in monospecific A. alba stands growing on eutrophic sites, the ratio between the number of seedlings aged 5–20 years and the seed production per m2 was estimated at about 0.05% (Paluch and Jastrzębski 2013), with major drivers of this phenomenon being fungal pathogens (Jankowiak et al. 2016). Their impact tends to intensify in wet years and in moist microhabitats associated with larger canopy gaps but also smaller inter-crown openings. This frequently leads to over-proportional seedling accumulation in the undercrown areas and absence of regeneration in inter-crown gaps (Paluch and Stepniewska 2012; Paluch et al. 2016). Nevertheless, juvenile natural regeneration of A. alba is characterized by a considerably higher survival rate in mixed stands than in monospecific stands, which suggests a higher intensity of environmental filtering in the latter case (Paluch and Jastrzębski 2013).
The strong environmental filtering at the stage of establishing regeneration and low parental tree density could potentially influence the gene flow between the parent and offspring populations. To test that conjecture, four hypotheses were formulated: (i) the genetic distance between parent and offspring populations increases with decreasing parent tree density, (ii) the genetic distance between parent and offspring populations differs in stands having different regeneration abundance, (iii) the SGS of the offspring populations in stands with a high adult tree density is weaker than in stands with a low adult tree density, and (iv) the SGS of the offspring populations differs in stands exhibiting different regeneration abundance.
Methods
Study region and sampling
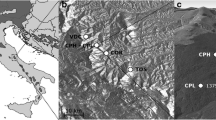
This study was conducted in the Western Carpathians (southern Poland) in stands located in the lower montane belt. In that region, depending on elevation, location, and aspect, the growth period (with a mean daily temperature above 5 °C) is ca. 180–200 days. The average annual temperature is 5–7 °C, and the annual precipitation is between 750 and 1450 mm with 60% occurring between May and October (Paszyński and Niedzwiedź 1999). In this area, A. alba occupies 26% of the total forested area. Growing on eutric, loamy cambisols developed on flysch material, it commonly occurs in mixed forests with Fagus sylvatica and/or Picea abies (Matuszkiewicz 2001). It also occurs in monospecific stands that originate from naturally established regeneration. However, these are mostly degraded forms of natural mixed forests which have been modified by the past selective exploitation of beech for fuel and charcoal, pasturing and litter raking practices, and silvicultural operations to promote conifers.
For this study, 17 stands with dominant A. alba were selected (hereafter, Abies stands) together with 10 stands with dominant Fagus sylvatica (hereafter, Fagus stands). The stands were located at an altitude between 500 and 950 m a.s.l. in an area defined by longitude 18.794°E and 22.733°E and latitude 49.133°N and 49.714°N (Online Resource 1, Table 1). Both species composition variants were studied in three to six stands in the eastern, middle, or western part of the Western Carpathian massif. The stands were characterized by the homogeneity of the site conditions and the stand structure, a lack of intensive silvicultural cutting or natural disturbances during the last 25 years, and the existence of a naturally established, multi-aged A. alba seedling bank consisting, according to the internodia counts, mostly of individuals aged under 25 years. The mature stands consisted of one tree layer and originated from natural regeneration in the shelterwood system with multiple removal cuttings over a 20–60 year period. The mean age of the stands ranged between 80 and 110 years, although the age of individual trees forming the stands varied.
More detailed characteristics of the parent stands and their regeneration were determined in 25 circular plots of radius 10 m, regularly spaced every 20 m in the core zone of each stand and are shown in Online Resource 1, Table 1. The density of adult A. alba trees with free tops and the potential to produce seeds (i.e., from Kraft’s classes 1–3) ranged between 153 and 517 per ha (mean = 280 per ha) in the Abies stands and between 57 and 194 per ha (mean = 111 per ha) in the Fagus stands. The differences in the density of adult A. alba trees resulted from the species composition, stand age, and past silvicultural treatments (light or heavy thinning in middle-aged stands). The visually estimated proportions of stand areas with A. alba seedling density above 2 individuals per m2 (hereafter, regeneration coverage) ranged between 6 and 58% (Online Resource 1, Table 1). To introduce a measure for environmental filtering (i.e., more or less favorable conditions for regeneration establishment), a variable calculated as the ratio of regeneration coverage and the density of adult A. alba trees per ha (hereafter, relative regeneration abundance) was used in the analyses. The density of adult A. alba trees per ha was treated here as an approximation for the quantity of seeds produced, and a higher or lower ratio of regeneration coverage per a unit of produced seeds was meant to reflect more or less favorable conditions for regeneration establishment. This measure was alternatively defined as the ratio of regeneration coverage to the basal area of adult A. alba trees per ha; however, both ratios were strongly correlated (r = 0.83) and produced similar results.
In the core zone of every stand, five reference points were selected: one midpoint and four points situated along the cardinal directions 30 m from the midpoint. For each of these five reference points, the six nearest A. alba trees growing in the main canopy were selected for genetic analyses (30 adult trees per stand, 810 in total). Within a 1–5 m radius of each of the adult trees, one A. alba seedling, aged approximately 5–25 years and below 0.7 m height, was randomly selected to make up a matching sample of 30 seedlings per stand (810 in total). The coordinates of the adult trees and the seedlings were determined accurate to 0.1 m using a precision compass and an ultrasonic distance meter. Later computations indicated that, depending on the stand species composition, the sampled area varied between 0.6 and 1.3 ha. Collecting seedlings aged less than 25 years (checked by an internodia count) prevented the sampling of progeny whose local parents had since been removed from the stand. In June 2016 a sample of the current year’s needles was taken from every adult tree and seedling, packed in plastic bags, labeled, and transported to the laboratory.
Molecular analysis
After the plant material was lyophilized in Labconco FreeZone 2.5 and pulverized in the Retsch MM400 mixer mill, a modified CTAB method was used to extract the genomic DNA from 20 mg of powdered dry needles (Khanuja et al. 1999). The resulting pellets were suspended in 50.0 µL TE buffer and ten times diluted. PCRs were then performed in a final volume of 15.0 µL containing a 3.0 µL DNA template, 1.5 µL 10 × DreamTaq Buffer, 2.0 mM MgCl2 (Thermo Fisher Scientific, USA), 0.2 mM of each dNTP (Thermo Fisher Scientific, USA), 0.5U DreamTaq™ DNA Polymerase (Thermo Fisher Scientific, USA), and 0.2 µM of primers F (labeled fluorescent) and R.
Preliminary seven most polymorphic nuclear microsatellite markers (of 11 nSSRs given in Cremer et al. 2006) were tested. Finally, for further analyses five nSSR markers were selected which provided most repeatable and high-quality amplification products: SFb4 (VIC TD65), SFb5 (NED TD65), SF78 (FAM TD65), SF239 (VIC TD68), SF333 (PET TD65). In a optimization step, two touchdown profiles, TD65 and TD68, were established and the amplification was then carried out in an Eppendorf Gradient S thermal cycler (Eppendorf AG, Germany) under the following conditions: 5 min at 95 °C followed by 10 touchdown cycles of 30 s at 95 °C, 30 s at 65 °C (TD65) or at 68 °C (TD68) (1 °C lower per cycle), 40 s at 72 °C and 25 cycles of 30 s at 95 °C, 50 s at 55 °C (TD65) or 58 °C (TD68), and 40 s at 72 °C with a final extension time of 8 min at 72 °C. Finally, the PCR products were separated by capillary electrophoresis using an automated sequencer 3500 Genetic Analyzer (Applied Biosystems, USA). The alleles were sized and analyzed using the internal size standard GeneScan™- 600 LIZ® Size Standard v2.0. (Applied Biosystems, USA) by the GeneMapper ® Software Version 4.0 (Applied Biosystems, USA). In the case of doubtful results, the PCR amplification and sizing was repeated to confirm the fragment lengths.
The nuclear microsatellite data were screened for genotyping errors, null alleles (non-amplifiable fragments), large-sized allele dropout, and stuttering using Micro-Checker v.2.2.3 (Van Oosterhout et al. 2004, 2006). In the analytic data set, individuals having more than one locus with no bands seen were discarded (28 individuals in total), but the set included those with only one such locus (133 in total). As a result, the data set included 1592 genotyped individuals. After estimating the null allele frequency in the single populations, the allele frequencies of the visible alleles have been adjusted using the method described in Brookfield (1996). Then, GENEPOP 3.4 (Raymond and Rousset 1995) was used to estimate deviations from the Hardy–Weinberg equilibrium, and the linkage disequilibrium for each locus pair and in each population, with the significance levels set at α = 0.05 and adjusted for multiple comparisons using the sequential Bonferroni correction. The test for linkage disequilibrium also indicated a lack of linkage between pairs of loci across all populations, although a limited number of loci in linkage disequilibrium were observed at a local level. The parameters of genetic variability were estimated using adjusted data corrected for null allele frequency (Van Oosterhout et al. 2006).
The genetic structure of parent versus offspring populations
For the adult tree and seedling populations in each of the studied stands, separate GenAlEx 6.5 computations (Peakall and Smouse 2006, 2012) were performed for the number of alleles per locus (Na), the number of rare alleles per locus (i.e., these with a frequency < 5% in a given population, Nr), the observed heterozygosity (Ho), the expected heterozygosity (He, unbiased estimate from Nei 1978), and the population inbreeding coefficient (FIS). These characteristics were compared for adult trees versus seedlings and the differences were given in percentages of the corresponding parent population values. The statistical significance of the differences between the adult trees and the seedlings in the individual stands and in the groups of Abies and Fagus stands were tested using permutation tests (10,000 permutations) in which genotypes were randomly assigned to adult trees or seedlings in each stand. The differences in genetic characteristics were then recalculated and compared with the results obtained from the original data. To account for multiple comparisons, a Holm–Bonferroni correction was used to assess the statistical significance of the differences between the empirical and simulated genetic parameters.
The short-term effective population size (Ne) was estimated from the adult tree and seedling populations using the estimator of standardized variance in the temporal changes of allele frequency developed by Nei and Tajima (1981). This quantity is defined as the size of an idealized Wright–Fisher population, which would give the same value of the variance of change in gene frequency resulting from a single generation of genetic sampling as in the population in question (Wang 2005). The genetic differentiation between adult and seedling cohorts in each population was expressed by a fixation index (FST) calculated using GenAlEx 6.5.
To check for possible shifts in allele frequencies at the local scale (i.e., in the individual stands), the genetic distances between the adult trees and the seedlings were calculated based on the shared allele distance (DSA, Bowcock et al. 1994), the Nei’s standard genetic distance (DS, Nei 1972), the arc distance (DA, Cavalli-Sforza and Edwards 1967), and the Prevosti distance (DP, Prevosti et al. 1975) (whose formulae are given in Online Resource 1, Table 2). In particular, the arc distance is robust to the presence of null alleles (Chapuis and Estoup 2007). The relationships between the density of adult A. alba trees and the regeneration coverage as explanatory variables and the short-term effective population size (Ne), the fixation index (FST), and the genetic distances between the adult trees and the seedlings (DSA, DS, DA, DP) as response variables were tested by correlation analysis. The statistical significance of the Pearson’s correlation coefficients was assessed using one-sided permutation tests (100,000 permutations).
Spatial genetic structure (SGS)
To express the correlation between the allele frequencies of individuals i and j, the relationship coefficient rij was computed as the correlation between individual allele frequencies (Hardy and Vekemans 2002). One advantage of the relationship coefficient is that it is not influenced by the selfing rate at constant gene flow parameters (Hardy and Vekemans 1999). To account for the presence of null alleles, the calculation procedure distinguished between the observed genotype and the set of true genotypes which may have produced the observed genotype, based on the estimated frequency of null alleles in the population (Wagner et al. 2006).
To examine the intensity and spatial range of SGS, the relationship coefficient rij for all i − j pairs was averaged for the distance intervals to yield mean values of Moran’s I statistic rij(d) widely used in ecological studies (Hardy and Vekemans 1999). The expected value of the relationship coefficient rij(d) for the case of the absence of spatial dependence is 0, with higher or lower values indicating that the individuals distanced by a distance d were more or less related than in a randomly drawn sample. To ensure a minimum sample size of 30 pairs in each distance interval (Jump and Peñuelas 2007), the coefficient was calculated using 10 m and 5 m wide distance intervals for individual stands and for the Abies and Fagus groups, respectively, with the analysis confined to distances less than 60 m. Here, rij(d) was calculated for adult trees, for seedlings, and for seedlings versus adult trees (cross-correlation) using the allele frequencies from the entire adult and seedling population as the reference sample. These calculations were performed separately for each stand and for the pooled Abies and Fagus stand groups.
Because an advection of air masses from the western and southern quadrant prevails in this region (Paszyński and Niedzwiedź 1999), the isotropy of the spatial correlation of seedlings versus adult trees was checked by calculating directional spectra in four directions (NE, SE, SW, and NW), as well as in the upslope and downslope directions. On the slopes, only adult tree-seedling pairs were considered when the slope between their locations was greater than 15% (otherwise the terrain was deemed to be plane). Moreover, the calculations included only 21 stands containing more than 10 such pairs in the first distance interval (< 5 m). Because of the reduced number of pairs, the directional spectra were calculated only for the total population of all stands.
The SGS intensity was expressed by the relationship coefficient rij calculated for distances below 10 m, in which the highest deviations from the random patterns were exhibited. The effect of the varying number of pairs in the individual stands was corrected by dividing the value of rij by half the 95% confidence interval width from the random labeling simulations. This transformation resulted in standardized values of > 1 or < − 1 for the relationship coefficient rij(d) values beyond the 95% confidence interval. The spatial range of SGS was assumed to be the distance at which the relationship coefficient rij(d) became 0, indicating a random distribution of the genotypes. This distance was determined as \({ \exp }\left( { - b/a} \right)\), where a and b are the slope and intercept of the regression between rij(d) and the logarithm of distance d as a covariate.
For better comparability of the results, the SGS was also expressed by the Sp statistics: \(S_{p} = - b/\left( {1 - \hat{\rho }_{ij} (1)} \right)\), in which \(\hat{\rho }_{ij}\) is the estimator of kinship coefficient (Loiselle et al. 1995), b is the regression slope of \(\hat{\rho }_{ij}\) on the logarithm of distance d, and \(\hat{\rho }_{ij} \left( 1 \right)\) denotes the kinship coefficient in the first distance interval. The Sp statistics is an integrated measure for comparing both the intensity and extent of SGS (Vekemans and Hardy 2004). The exact tests for the statistical significance of the Sp statistics were obtained by comparing the empirical Sp values with the values from the random labeling simulations (10,000 randomizations for each stand).
The intensity and spatial range of SGS were compared for stands with different densities of adult trees and seedling abundance. Statistical significances of the differences between the Abies and Fagus stands were tested by bootstrapping the residuals of the respective regression models (two-sided tests, 10,000 bootstrap samples) (Efron and Tibshirani 1993, pp. 111–115). Correlation analysis was then used to compare the SGS in the stands having different characteristics, and the statistical significance of the Pearson correlation coefficients was tested using permutation tests (100,000 permutations). The analysis of the SGS was programmed in Visual Basic for Applications language.
Results
Shifts in the stand level characteristics
A total of 148 alleles were identified in the entire population of 1592 individuals: 75, 33, 17, 15 and 8 at the loci SF78, SFb4, SFb5, SF239 and SF333, respectively. The average null allele frequencies were 0.01, 0.05, 0.19, 0.19 and 0.20, respectively. The frequency of null alleles did not vary significantly between the Abies and Fagus stands, or between the seedling and adult tree populations (data not shown). Three loci with high null allele frequencies (i.e., SF239, SFb5, and SF333) showed a significant excess of homozygotes and inbreeding coefficients (FIS) between and 0.309 and 0.491 (Online Resource 1, Table 3).
In the adult tree populations, the mean number of alleles per locus (Na) in the individual stands varied between 7.8 and 12.2 (with a mean for all stands of 10.0, Table 1). In the total population, however, 50% of the alleles occurred at a frequency of less than 5%, and in the individual stands the percentage of rare alleles (i.e., these with a frequency < 5% in a given population, Nr) varied between 40 and 62%. The observed heterozygosity (Ho) ranged between 0.429 and 0.653, and the expected heterozygosity (He) between 0.660 and 0.796. The average inbreeding coefficient (FIS) was 0.229 in the total adult tree population, and this value varied between 0.168 and 0.459 in the individual stands.
The seedling populations on average contained fewer alleles than the adult tree populations: −1.7% lower in the Abies stands, −4.1% in the Fagus stands, and −2.6% in the entire population (Table 1). Nevertheless, according to the permutation tests, in neither case were the differences statistically significant at the conventional 0.05 level. In the individual stands, in contrast, both decreasing (up to −15.5%) and increasing (up to + 28.2%) tendencies were observable, with the changes being statistically significant in three stands, two of which showed a decreasing Na in the offspring populations. No significant correlation emerged, however, between the shifts in the number of alleles per locus (Na) and the density of adult trees, the regeneration coverage, or the relative regeneration abundance.
The analysis did reveal substantial changes in the number of rare alleles (Nr) between the adult tree populations and the regeneration, ranging widely from −50.0% to + 72.2% in the individual stands (Table 1). Although the decrease was stronger in the Fagus (−14.3%) than in the Abies stands (−0.5%), the difference between these groups was not statistically significant. The correlation analysis led to the same conclusion: the shifts in the number of rare alleles (Nr) did not correlate with the density of adult trees. Nor did any significant correlation emerge between these shifts and the regeneration coverage or the relative regeneration abundance. According to the permutation tests, none of the shifts in the number of rare alleles (Nr) in either the Fagus, the Abies, or the combined population was significant. Likewise, in the pooled Abies and Fagus stands, no significant shifts in either the observed (Ho) or the expected heterozygosity (He) were evident between the adult tree and seedling populations. Two Fagus stands and one Abies stand were characterized by a significant reduction in the observed heterozygosity (Table 1).
As indicated by the FST index (corrected for null alleles), in the total pool of all stands the genetic differentiation between adult and seedling populations was 0.04% and did not reach statistical significance (data not shown). In the individual stands, this genetic differentiation varied between 0.5 and 3.3% and was statistically significant in five stands (Table 2). The proportion was higher in the Fagus than in the Abies stands and tended to increase as the density of adult trees decreased (r = − 0.52, p < 0.001, Fig. 1a). Considering this effect, the partial correlation coefficients between the FST values and the relative regeneration abundance and between the FST values and the regeneration coverage were insignificant. The shared allele distance, standard Nei distance, arc distance, and Prevosti distance (Fig. 1b) increased with decreasing density of adult trees (r = − 0.25, p = 0.08, r = − 0.47, p < 0.001, r = − 0.33, p = 0.44; and r = − 0.35, p = 0.04, respectively). However, no meaningful correlation emerged between these distances and the regeneration coverage or the relative regeneration abundance.
Genetic differentiation and Prevosti genetic distance between adult and seedling generations (a and b, respectively), and short-term effective population size (c) determined in the 27 study stands with different adult tree densities. In each stand the analyses were carried out for 30 adult trees and 30 seedlings based on five microsatellite markers. Comment: The genetic differentiation and genetic distance between adult trees and seedlings tend to increase as the density of adult trees decreases
The short-term effective population size (Ne) varied between 7.4 and 42.3 and was significantly higher in the Abies stands (25.7) than in the Fagus stands (14.2) (p = 0.009). A significant correlation was found between the effective population size (Ne) and the density of adult trees (r = 0.51, p < 0.001) (Fig. 1c) but not between Ne and either the regeneration coverage (r = 0.31, p = 0.12), or the relative regeneration abundance (r = − 0.20, p = 0.19). These results indicate that with an adult tree density of about 50 individuals per ha, Ne attains the level of about 13 individuals (Fig. 1c).
Spatial genetic structure (SGS)
The SGS of adult trees analyzed across all the Abies and Fagus stands differed significantly (Fig. 2). Whereas the rij(d) values for the former fell within the 95% confidence interval for random patterns over the entire distance range analyzed, the values for the latter departed significantly from randomness (Sp = 0.0, p = 0.002) with a positive correlation detectable in a distance range below 30.4 m.
Spatial genetic correlation in the adult A. alba populations depending on distance: the empirical means of the relationship coefficient rij in 5 m width distance intervals ± SE (points and whiskers) and 95% confidence intervals obtained from random labeling simulations (dashed lines). The empirical values beyond the confidence intervals indicate a significant spatial genetic structure. The significance of the Sp statistics, being a measure of intensity and extent (spatial scale) of the genetic correlation, was tested by comparing the empirical values with the values obtained from random labeling simulations (10,000 randomizations for each stand). a All stands pooled. bAbies stands. cFagus stands. Comment: The spatial genetic structuring is evident in the Fagus stands up to a distance of 30.4 m but not in the Abies stands
In the pooled sample for seedlings (from all 27 stands), the SGS was significant for distances below 28.0 m (Sp = 0.010, p = 0.03) (Fig. 3). However, the correlograms did not attain the conventional significance level when the Abies (Sp = 0.009, p = 0.08) and the Fagus stands (Sp = 0.011, p = 0.11) were analyzed separately. Moreover, the spatial range of the genetic correlation, however, did not statistically differ between the Abies and Fagus stands (p > 0.2).
Spatial genetic correlation in the A. alba seedling populations: the empirical means of the relationship coefficient rij in 5 m width distance intervals ± SE (points and whiskers) and 95% confidence intervals obtained from random labeling simulations (dashed lines). a All stands pooled. bAbies stands. cFagus stands. Comment: In the pooled seedling sample for all stands, spatial genetic structure is discernible and statistically significant up to a distance of 28.0 m. Because of smaller sample size, in the Abies and Fagus stands the statistical significance of the correlograms is lower, but the general tendencies are similar as in the pooled sample
The genotypes of adult trees and seedlings showed positive spatial cross-correlation in both the Abies (Sp = 0.010, p < 0.001) and the Fagus stands (Sp = 0.013, p < 0.001) (Fig. 4). In the entire population, the spatial range of the genetic cross-correlation between seedlings and adult trees was 22.4 m. This characteristic did not statistically differ in the Abies and the Fagus stands (p > 0.2). The Sp statistics for the cardinal directions varied between 0.004 and 0.011, while those for the upslope and downslope directions were 0.006 and 0.007, respectively. In neither case were the differences between the directional spectra statistically significant.
Spatial genetic cross-correlation between the populations of seedlings and adult trees: the empirical means of the relationship coefficient rij in 5 m width distance intervals ± SE (points and whiskers) and 95% confidence intervals obtained from random labeling simulations (dashed lines). a All stands pooled. bAbies stands. cFagus stands. Comment: The genotypes of adult trees and seedlings showed positive spatial cross-correlation in the Abies and Fagus stands and the spatial range of this correlation was similar (22.4 m)
The SGS intensities tended to increase and the SGS extent to decrease as the number of A. alba decreased in the parent stands. This trend was clearly visible in the adult tree populations (r = − 0.40, p = 0.03, Fig. 5a) and discernible in the seedling populations (r = − 0.30, p = 0.09, Fig. 5c). Similarly, in the adult tree and seedling cohorts the range of SGS tended to decrease as the number of A. alba decreased in the parent stands (Fig. 5b, d). The intensity of genetic correlation increased and its spatial extent decreased as the regeneration coverage decreased (r = − 0.30, p = 0.02, Fig. 6), but both these variables remained uncorrelated with the relative regeneration abundance. No significant correlation was found between the SGS intensity of the adult trees and the SGS intensity of the seedlings.
Relationship between the density of adult A. alba trees and the intensity and range of spatial genetic correlation in the adult tree (a, b) and seedling (c, d) populations. The intensity of spatial correlation is expressed by the relationship coefficient rij calculated for distances below 10 m and standardized through division by half the 95% confidence interval width obtained from the random labeling simulations. The standardized values of rij> 1 indicate significant positive genetic correlations. Comment: In the adult tree and seedling populations the spatial genetic correlation deepen both in the terms of intensity and extent as the number of adult trees decreases
Relationship between the regeneration coverage in the individual stands and the spatial genetic correlation in the seedling populations. The degree of spatial correlation is expressed by the relationship coefficient rij calculated as described in Fig. 5. Comment: The genetic correlation is stronger and spatially more extensive in the stands with lower regeneration density
Discussion
Regenerative processes in forest ecosystems are highly stochastic because of variation in annual seed crop, seed dispersal, site heterogeneity and the impact of biotic factors affecting seed and seedling survival. Therefore, before making generalizations about the mechanisms driving forest regeneration and the processes involved extensive sampling to capture their spatiotemporal variation is essential. Jump et al. (2012) demonstrated that the extent of SGS can vary by an order of magnitude within the same species because of different molecular markers, inter-plot variation, management history, and/or sampling regime. For that reason, in the current study more effort was put into sampling more stands than increasing the number of markers and individuals in the single locations. The results confirm that even under similar site and stand conditions considerable variation in gene flow and the resulting SGS can be expected. Although the sample of 27 stands studied seems representative to pinpoint the tendencies at the meta-population level, one limitation of the sampling scheme used remains its lower power at the within-stand level. In fact, the single study populations were smaller than simulations have suggested is ideal for SGS analyses. Cavers et al. 2005 showed that for 5 microsatellite markers a mean correlation between real and sampled distogram of the spatial genetic structure close to 0.9 is achievable with a sample of 100 individuals (and a higher number of individuals for high gene flow species). Nevertheless, in almost all the stands studied here, the analysis included the recommended > 30 pairwise comparisons per distance class (Jump and Peñuelas 2007).
Another problem is that three of the five SSRs used in this study showed a considerable number of null alleles. The presence of null alleles may be particularly problematic in studies comparing different sets of populations with different frequencies of null alleles and/or patterns of gene flow, especially when one or several population sets are characterized by low levels of gene flow (Chapuis and Estoup 2007). However, it seems that this may not significantly affect the SGS found in this study since the frequencies of the null alleles were similar in the adult tree and seedling populations and most probably independent from the spatial distribution of the genotypes. In addition, to accommodate for the presence of null alleles, the calculation procedure distinguished between an observed genotype and the set of true genotypes that may have produced that observation given an estimated frequency of null alleles in the population. Following the procedure described by Wagner et al. (2006) which improves estimates compared with ignoring the presence of null alleles or discarding the affected loci, for each two individuals the likelihood of their hypothetical true genotypes were compared for candidate relationships (full-sibs, half-sibs, first cousins or unrelated for the individuals from the same generation, and parent–offspring or unrelated in intergenerational comparisons), and then the genotypic data with the maximum likelihood were used to calculate the relationship coefficient.
Genetic distance between adult trees and seedlings
In the analyzed gradient of decreasing adult tree density, a tendency toward an increased genetic distance (or decreased genetic similarity) between the parent and offspring populations was observed. A decreased adult tree density was also associated with increased FST values (i.e., genetic differentiation between the adult and offspring populations) and a reduced effective population size. These alarming tendencies, however, were not observable in cumulative measures such as the number of alleles or the heterozygosity. Moreover, although the intergenerational (adult vs. seedling) differences in allele numbers or heterozygosity reached high values in the individual stands, they were statistically significant only in few cases. The findings of this study thus echo earlier research reporting only slight intergenerational changes in genetic structure for A. alba, Larix decidua Mill., Picea abies, Pinus mugo Turra (Müller-Starck et al. 2000), Abies amabilis Douglas ex J. Forbes, Tsuga heterophylla (Raf.) Sarg. (El-Kassaby et al. 2003), and Picea glauca (Moench) Voss (Rajora 1999). Several studies have demonstrated that neither variations in stand density nor different cutting methods lead to significant changes in the mating systems of wind-pollinated tree species (Neale and Adams 1985; El-Kassaby and Jaquish 1996; Stoehr 2000; Perry and Bousquet 2001).
The analysis identified no statistically meaningful associations between the regeneration abundance and the genetic differentiation between parents and their offspring. This result may suggest that in the stands under study the availability of A. alba seeds and environmental filtering were randomly coupled in space and time and did not affect the offspring populations’ genetic diversity. Nonetheless, the lack of significant differences in allele number and heterozygosity, concurrent with the trend in which the genetic distances between parent trees and seedlings increased as the population density decreased, may suggest some compensating mechanisms (e.g., increasing immigration rates) when there are fewer adult trees.
Spatial genetic structure (SGS)
One integrated measure for comparing both the intensity and extent of a SGS is the Sp statistic. Vekemans and Hardy (2004) reported average Sp values of 0.0102 for trees, and a variation between 0.0020 and 0.0108 across five wind-pollinated tree species. In this current study the Sp statistic was estimated at about 0.007 for seedlings and 0.019 for adult trees in the Fagus stands. In general, family structures tend to be stronger in the younger than in the older cohorts, possibly because of historical factors (bottlenecks or founder effects), local adaptation due to microhabitat selection, or changes in the population density with maturation (Sagnard et al. 2011). Not only can this effect probably explain the lack of SGS in adult trees in the Abies stands, but it suggests that the stronger SGS in adult trees relative to seedlings in the Fagus stands may be the result of single reproductive events involving only a few trees of the previous generation (Jones and Hubbell 2006; Troupin et al. 2006). This assumption is plausible because, despite the lack of historical data to directly support it, strong fluctuations in species composition across the generations are common in forests with A. alba and Fagus sylvatica (Paluch and Jastrzębski 2013).
The spatial range of genetic correlation shows considerable variation between tree species because of their different pollen and seed dispersal characteristics. For wind-pollinated forest tree species, the extent of a SGS is generally accepted to be limited to 30–50 m in Fagus sylvatica and Quercus petraea (Matt.) Liebl. and up to 60 m in Quercus robur L. and Fraxinus excelsior L. (Jump et al. 2012). In the current study, the spatial range of the correlation between adult trees and seedlings was estimated at 22.4 m and that between seedlings at 28.0 m. One important source of the genetic structure observed at the shortest distances is most probably maternal half-sibs growing in close proximity to their source trees. This assumption is supported by the good fit between the estimated range of the spatial correlation and the reported dispersal distances of A. alba seeds in forest interiors. Depending on the stand density, the median seed dispersal distance has been estimated at between 6 and 21.5 m, with the maximum density of fallen seeds per unit area found at a distance range of 3–6 m from the mother tree (Sagnard et al. 2007; Amm et al. 2012; De Andrés et al. 2014).
As anticipated, and as evidenced elsewhere (Vekemans and Hardy 2004; Kyndt et al. 2009), the SGS intensity tended to increase as the density of adult trees decreased. It may be assumed that, when the adult tree density is high, the microhabitats may be supplied by multiple seed and pollen donors, what may reduce the likelihood of SGS being established (Nakanishi et al. 2005). A simulation study by Sagnard et al. (2011) demonstrated that, given the dispersal function parameters realistic for A. alba, a SGS appeared with an adult tree density of less than 9 per ha. The model in that simulation, however, did not include cumulative reproductive events, the stochasticity of flower phenology, seed production, or germination over time. In this study, carried out in the Western Carpathians, of particular note is the occurrence of spatial structures in stands characterized by a relatively high density of parent trees and probably strong overlap of their seed and pollen shadows. The results obtained suggest that the SGS may be expected at densities below 130 individuals per ha in the adult tree populations (see the values of rij > 1 on Fig. 5a), and even at densities below 200 individuals per ha in the seedling populations (Fig. 5c).
The significant SGS identified in the stands with a high density of potential parent trees may be linked with a higher fecundity of some individuals, or the coincidence of their crop year, or years, with advantageous environmental conditions. Indeed, some studies have found a higher across-year variation in seed production than across the sampling sites (Beckage et al. 2005). Similarly, a simulation study by Paluch (2011) demonstrated that even under conditions of strongly overlapping seed shadows, linking the fecundity rate with the basal area of the mother trees is sufficient to evoke the effect of a strong spatial dependence of seed density in the ground. Moreover, the canopy density may be a more important determinant of the mating system than the within-population mate density (Gauzere et al. 2013) because shorter dispersal distances are expected in dense vegetation (Guries and Nordheim 1984; Nathan et al. 2011). Compared to other conifers, A. alba produces very heavy pollen of a sedimentation velocity of ca. 0.12 m s−1 and a weight of ca. 251.1 × 10−6 g per 1000 grains (Eisenhut 1961) and the vast majority of the pollen is deposited within less than 50 m of the sampling site and a major proportion within 100 m (Poska and Pidek 2010). Restoux (2009) reported a mean dispersal distance of less than 10 m for A. alba pollen in a 70-year-old stand dominated by spruce. Therefore, when the canopies are closed, the effective bounds of gene flow may be quite limited, with few major donors and numerous minor contributors (Kang et al. 2003; Robledo-Arnuncio et al. 2004b; Nijensohn et al. 2005).
Conclusions
Even though the identified shifts were small, the results corroborate the expectation that a decreasing population density is accompanied by the decreasing genetic similarity between the parents and their progeny. However, there was no evidence that regeneration abundance had any influence on intergenerational gene flow, suggesting that in the stands studied the availability of seeds and the environmental filtering were randomly linked in time and space. This study showed that in the individual stands the SGS varied in both its spatial extent and its intensity, and that the SGS in the younger generation tended to deepen as the population density and regeneration abundance decreased. There was no tendency, however, for the SGS intensity to deepen in the offspring population relative to adult trees, which may suggest that the effect of the initial SGS in the parent populations can be overridden by other concomitant factors.
With regard to strategies for management and genetic conservation practices, the study results emphasize that in mixed-species stands with a low density of A. alba trees (below ca. 50 individuals per ha) there is an alarming reduction in the effective population size (below ca. 13 individuals) and a risk of disturbed vertical gene flow resulting from the stochastic sampling a finite number of gametes during reproduction and survival. To increase the number of mating individuals, diminish founder effects and reduce risk of inbreeding, in such stands the period of regeneration establishment should last possibly long, and selective cuttings inherently reducing the number and distances between the potential parent trees should be avoided. Moreover, preferring regeneration established in the stand patches influenced by overlapping seed/pollen shadows of several parents against cohorts of maternal half-sibs growing in the close vicinity of solitary trees may decrease relatedness among the neighboring individuals and increase genetic diversity in the offspring population.
This study has shown that in the stands with a number of individuals less than 130 per ha, a significant spatial correlation of the genotypes in a range up to ca. 40 m may be expected (Fig. 5a, b). The SGS has consequences for the seed collection practices, because the genetic diversity of the seed lots is strongly affected by the number of trees designated for the seed harvest and their spatial arrangement (Konnert and Hosius 2010). Therefore, for seed collection uniform harvesting across the whole stand area with a minimum distance of about 40 m between the trees designated for the seed harvest might be recommended.
References
Amm A, Pichot C, Dreyfus P, Davi H, Fady B (2012) Improving the estimation of landscape scale seed dispersal by integrating seedling recruitment. Ann For Sci 69:845–856. https://doi.org/10.1007/s13595-012-0208-1
Beaudet M, Brisson J, Gravel D, Messier C (2007) Effect of a major canopy disturbance on the coexistence of Acer saccharum and Fagus grandifolia in the understorey of an old growth forest. J Ecol 95:458–467. https://doi.org/10.1111/j.1365-2745.2007.01219.x
Beckage B, Lavine M, Clark JS (2005) Survival of tree seedlings across space and time: estimates from long-term count data. J Ecol 93:1177–1184. https://doi.org/10.1111/j.1365-2745.2005.01053.x
Bladé C, Vallejo VR (2008) Seed mass effects on performance of Pinus halepensis Mill. seedlings sown after fire. For Ecol Manage 255:2362–2372. https://doi.org/10.1016/j.foreco.2007.12.039
Bowcock AM, Ruiz-Linares A, Tomfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457. https://doi.org/10.1038/368455a0
Breed MF, Marklund MH, Ottewell KM, Gardner MG, Harris JBC, Lowe AJ (2012) Pollen diversity matters: revealing the neglected effect of pollen diversity on fitness in fragmented landscapes. Mol Ecol 21:5955–5968. https://doi.org/10.1111/mec.12056
Brookfield JF (1996) A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol 5:453–545. https://doi.org/10.1046/j.1365-294X.1996.00098.x
Buck JR, Clair SBS (2014) Stand composition, proximity to overstory trees and gradients of soil moisture influence patterns of subalpine fir seedling emergence and survival. Plant Soil 381:61–70. https://doi.org/10.1007/s11104-014-2094-9
Cavalli-Sforza LL, Edwards AWF (1967) Phylogenetic analysis: models and estimation procedures. Am J Hum Gen 19:233–257
Cavers S, Degen B, Caron H, Lemes MR, Margis R, Salgueiro F, Lowe AJ (2005) Optimal sampling strategy for estimation of spatial genetic structure in tree populations. Heredity 95:281–289. https://doi.org/10.1038/sj.hdy.6800709
Chapuis MP, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631. https://doi.org/10.1093/molbev/msl191
Cremer E, Liepelt S, Sebastiani F, Buonamici A, Michalczyk IM, Ziegenhagen B, Vendramin GG (2006) Identification and characterization of nuclear microsatellite loci in Abies alba Mill. Mol Ecol Notes 6:374–376. https://doi.org/10.1111/j.1471-8286.2005.01238.x
De Andrés EG, Camarero JJ, Martínez I, Coll L (2014) Uncoupled spatiotemporal patterns of seed dispersal and regeneration in Pyrenean silver fir populations. For Ecol Manage 319:18–28. https://doi.org/10.1016/j.foreco.2014.01.050
De-Lucas AI, González-Martínez S, Vendramin G, Hidalgo E, Heuertz M (2009) Spatial genetic structure in continuous and fragmented populations of Pinus pinaster Aiton. Mol Ecol 18:4564–4576. https://doi.org/10.1111/j.1365-294X.2009.04372.x
Efron B, Tibshirani RJ (1993) An introduction to the bootstrap. Chapman and Hall/CRC, New York
Eisenhut G (1961) Untersuchungen über die Morphologie und Ökologie der Pollenkörner heimischer und fremdländischer Waldbäume. Forstwiss Forsch 15:1–68
El-Kassaby YA, Jaquish B (1996) Population density and mating pattern in western larch. J Heredity 87:438–443. https://doi.org/10.1093/oxfordjournals.jhered.a023034
El-Kassaby YA, Dunsworth BG, Krakowski J (2003) Genetic evaluation of alternative silvicultural systems in coastal montane forests: western hemlock and amabilis fir. Theor Appl Genet 107:598–610. https://doi.org/10.1007/s00122-003-1291-3
Finkeldey R, Ziehe M (2004) Genetic implications of silvicultural regimes. For Ecol Manage 197:231–244. https://doi.org/10.1016/j.foreco.2004.05.036
Gauzere J, Klein EK, Oddou-Muratorio S (2013) Ecological determinants of mating system within and between three Fagus sylvatica populations along an elevational gradient. Mol Ecol 22:5001–5015. https://doi.org/10.1111/mec.12435
Geburek T, Müller F (2005) How can silvicultural management contribute to genetic conservation? In: Geburek T, Turok J (eds) Conservation and management of forest genetic resources in Europe. Arbora Publishers, Zvolen, pp 651–669
Gérard PR, Klein EK, Austerlitz F, Fernández-Manjarrés JF, Frascaria-Lacoste N (2006) Assortative mating and differential male mating success in an ash hybrid zone population. BMC Evol Biol 6:96. https://doi.org/10.1186/1471-2148-6-96
Guries RP, Nordheim EV (1984) Flight characteristics and dispersal potential of maple samaras. For Sci 30:434–440
Hamrick JL, Murawski DA, Nason JD (1993) The influence of seed dispersal mechanisms on the genetic structure of tropical tree populations. In: Fleming TH, Estrada A (eds) Frugivory and seed dispersal: ecological and evolutionary aspects. Kluwer, Dordrecht, pp 281–298
Hardy OJ, Vekemans X (1999) Isolation by distance in a continuous population: reconciliation between spatial autocorrelation analysis and population genetics models. Heredity 83:145–154. https://doi.org/10.1046/j.1365-2540.1999.00558.x
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hoban SM, McCleary TS, Schlarbaum SE, Romero-Severson J (2014) Spatial genetic structure in 21 populations of butternut, a temperate forest tree (Juglans cinerea L.), is correlated to spatial arrangement, habitat, and land-use history. For Ecol Manage 314:50–58. https://doi.org/10.1016/j.foreco.2013.11.001
Hosius B, Leinemann L, Konnert M, Bergmann F (2006) Genetic aspects of forestry in the Central Europe. Eur J For Res 125:407–417. https://doi.org/10.1007/s10342-006-0136-4
Jankowiak R, Bilański P, Paluch J, Kołodziej Z (2016) Fungi associated with dieback of Abies alba seedlings in naturally regenerating forest ecosystems. Fungal Ecol 24:61–69. https://doi.org/10.1016/j.funeco.2016.08.013
Jones FA, Hubbell SP (2006) Demographic spatial genetic structure of the neotropical tree, Jacaranda copaia. Mol Ecol 15:3205–3217. https://doi.org/10.1111/j.1365-294X.2006.03023.x
Jump AS, Peñuelas J (2007) Extensive spatial genetic structure revealed by AFLP but not SSR molecular markers in the wind-pollinated tree, Fagus sylvatica. Mol Ecol 16:925–936. https://doi.org/10.1111/j.1365-294X.2006.03203.x
Jump AS, Rico L, Coll M, Peñuelas J (2012) Wide variation in spatial genetic structure between natural populations of the European beech (Fagus sylvatica) and its implications for SGS comparability. Heredity 108:633–639. https://doi.org/10.1038/hdy.2012.1
Kang KS, Bila AD, Harju AM, Lindgren D (2003) Estimation of fertility variation in forest tree population. Forestry 76:329–344. https://doi.org/10.1093/forestry/76.3.329
Khanuja S, Shasany A, Darokar M, Kumar S (1999) Rapid isolation of DNA from dry and fresh samples of plants producing large amounts of secondary metabolites and essential oils. Plant Mol Biol Rep 17:1–7. https://doi.org/10.1023/A:1007528101452
Konnert M, Hosius B (2010) Beitrag der Forstgenetik für ein nachhaltiges Waldmanagement. Forstarchiv 81:170–174. https://doi.org/10.2376/0300-4112-81-170
Konnert M, Hussendörfer E (2001) Genetic variation of silver fir (Abies alba) in uneven aged forests (”Plenter” forest) in comparison with even-aged forests (Altersklassenwald). In: Müller-Starck G, Schubert R (eds) Genetic response of forest systems to changing environmental conditions. Kluwer, Dordrecht, pp 307–320
Kyndt T, Assogbadjo AE, Hardy OJ, Glele Kakai R, Sinsin B, Van Damme P, Gheysen G (2009) Spatial genetic structuring of baobab (Adansonia digitata, Malvaceae) in the traditional agroforestry systems of West Africa. Am J Bot 96:950–957. https://doi.org/10.3732/ajb.0800266
Leonardi S, Piovani P, Scalfi M, Piotti A, Giannini R, Menozzi P (2012) Effect of habitat fragmentation on the genetic diversity and structure of peripheral populations of beech in central Italy. J Hered 103:408–417. https://doi.org/10.1093/jhered/ess004
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understorey shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Madsen P, Hahn K (2008) Natural regeneration in a beech-dominated forest managed by close-to-nature principles—a gap cutting based experiment. Can J For Res 38:1716–1729. https://doi.org/10.1139/X08-026
Matuszkiewicz JM (2001) Zespoły leśne Polski. Wydawnictwo Naukowe PWN, Warsaw
McClure JW, Lee TD, Leak WB (2000) Gap capture in northern hardwoods: patterns of establishment and height growth in four species. For Ecol Manage 127:181–189. https://doi.org/10.1016/s0378-1127(99)00129-2
Müller-Starck G, Konnert M, Hussendörfer E (2000) Empfehlungen zur genetisch nachhaltigenWaldbewirtschaftung—Beispiele aus dem Gebirgswald. For Snow Landsc Res 75:29–50
Nakanishi A, Tomaru N, Yoshimaru H, Manabe T, Yamamoto S (2005) Interannual genetic heterogeneity of pollen pools accepted by Quercus salicina individuals. Mol Ecol 14:4469–4478. https://doi.org/10.1111/j.1365-294X.2005.02736.x
Nathan R, Katul GG, Bohrer G, Kuparinen A, Soons MB, Thompson SE, Trakhtenbrot A, Horn S (2011) Mechanistic models of seed dispersal by wind. Theor Ecol 4:113–132. https://doi.org/10.1007/s12080-011-0115-3
Neale DB, Adams WT (1985) The mating system in natural shelterwood stands of Douglas fir. Theor Appl Genet 71:201–207
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nei M, Tajima F (1981) Genetic drift and estimation of effective population size. Genetics 98:625–640
Nijensohn SE, Schaberg PG, Hawley GJ, de Hayes DH (2005) Genetic subpopulation structuring and its implications in a mature eastern white pine stand. Can J For Res 35:1041–1052. https://doi.org/10.1139/x05-029
Nopp-Mayr U, Kempter I, Muralt G, Gratzer G (2012) Seed survival on experimental dishes in a central European old-growth mixed-species forest: effects of predator guilds, tree masting and small mammal population dynamics. Oikos 121:337–346. https://doi.org/10.1111/j.1600-0706.2011.19099.x
Paluch J (2005) Spatial distribution of regeneration in West Carpathian uneven-aged silver fir forests. Eur J For Res 124:47–54. https://doi.org/10.1007/s10342-005-0054-x
Paluch J (2011) Ground seed density patterns under conditions of strongly overlapping seed shadows in Abies alba Mill. stands. Eur J For Res 130:1009–1022. https://doi.org/10.1007/s10342-011-0486-4
Paluch J, Jastrzębski R (2013) Natural regeneration of shade-tolerant Abies alba Mill. in gradients of stand species compositions: limitation by seed availability or safe microsites? For Ecol Manage 307:322–332. https://doi.org/10.1016/j.foreco.2013.06.035
Paluch J, Stępniewska H (2012) Effect of microsites on the survival, density, and ectomycorrhizal status of shade-tolerant Abies alba regeneration attacked by fungal pathogens. Can J For Res 42:720–732. https://doi.org/10.1139/x2012-015
Paluch J, Kołodziej Z, Skrzyszewski J, Bartkowicz L, Gruba P (2016) Regeneration patterns of the late-successional Abies alba Mill.: inhibition in monospecific stands and colonization in mixed stands. Ann For Sci 73:1015–1024. https://doi.org/10.1007/s13595-016-0573-2
Paszyński J, Niedzwiedź T (1999) Klimat. In: Starkel L (ed) Geografia Polski. Środowisko przyrodnicze. PWN, Warsaw, pp 288–343
Peakall R, Smouse PE (2006) GenAlEx 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Perry DJ, Bousquet J (2001) Genetic diversity and mating system of post-fire and post-harvest black spruce: an investigation using codominant sequence-tagged site (STS) markers. Can J Forest Res 31:32–40. https://doi.org/10.1139/x00-137
Poska A, Pidek IA (2010) Pollen dispersal and deposition characteristics of Abies alba, Fagus sylvatica and Pinus sylvestris, Roztocze region (SE Poland). Veget Hist Archaeobot 19:91–101. https://doi.org/10.1007/s00334-009-0230-x
Prevosti A, Ocana J, Alonso G (1975) Distances between populations of Drosophila subobscura based on chromosome arrangement frequencies. Theor Appl Genet 45:231–241
Rajendra KC, Seifert S, Prinz K, Gailing O, Finkeldey R (2014) Subtle human impacts on neutral genetic diversity and spatial patterns of genetic variation in European beech (Fagus sylvatica). For Ecol Manage 319:138–149. https://doi.org/10.1016/j.foreco.2014.02.003
Rajora OP (1999) Genetic biodiversity impacts of silvicultural practices and phenotypic selection in white spruce. Theor Appl Genet 99:954–961
Raymond M, Rousset F (1995) GENEPOP (version 3.4): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Restoux G (2009) Variabilité spatio-temporelle de la reproduction chez une espèce pérenne monoïque: Le cas du sapin pectiné, Abies alba Miller, en limite sud de son aire de répartition. PhD thesis, University of Aix-Marseille
Restoux G, Silva DE, Sagnard F, Torre F, Klein E, Fady B (2008) Life at the margin: the mating system of Mediterranean conifers. Web Ecol 8:94–102. https://doi.org/10.5194/we-8-94-2008
Robledo-Arnuncio JJ, Alia R, Gil L (2004a) Increased selfing and correlated paternity in a small population of a predominantly outcrossing conifer, Pinus sylvestris. Mol Ecol 13:2567–2577. https://doi.org/10.1111/j.1365-294X.2004.02251.x
Robledo-Arnuncio JJ, Smouse PE, Gil L, Alia R (2004b) Pollen movement under alternative silvicultural practices in native populations of Scots pine (Pinus sylvestris L.) in central Spain. For Ecol Manage 197:243–253. https://doi.org/10.1016/j.foreco.2004.05.016
Sagnard F, Pichot C, Dreyfus P, Jordano P, Fady B (2007) Modelling seed dispersal to predict seedling recruitment: recolonization dynamics in a plantation forest. Ecol Model 203:464–474. https://doi.org/10.1016/j.ecolmodel.2006.12.008
Sagnard F, Oddou-Muratorio S, Pichot C, Vendramin GG, Fady B (2011) Effects of seed dispersal, adult tree and seedling density on the spatial genetic structure of regeneration at fine temporal and spatial scales. Tree Genet Genomes 7:37–48. https://doi.org/10.1007/s11295-010-0313-y
Snyder RE, Chesson P (2004) How the spatial scales of dispersal, competition, and environmental heterogeneity interact to affect co-existence. Am Nat 164:633–650. https://doi.org/10.1086/424969
Sork VL, Davis FW, Smouse PE, Apsit VJ, Dyer RJ, Fernandez-M JF, Kuhn B (2002) Pollen movement in declining populations of California valley oak, Quercus lobata: where have all the fathers gone? Mol Ecol 11:1657–1668
Stoehr MU (2000) Seed production of western larch in seed-tree systems in the southern interior of British Columbia. For Ecol Manage 130:7–15
Troupin D, Nathan R, Vendramin GG (2006) Analysis of spatial genetic structure in an expanding Pinus halepensis population reveals development of fine-scale genetic clustering over time. Mol Ecol 15:3617–3630. https://doi.org/10.1111/mec.12435
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x
Van Oosterhout C, Weetman D, Hutchinson WF (2006) Estimation and adjustment of microsatellite null alleles in nonequilibrium populations. Mol Ecol Notes 6:255–256. https://doi.org/10.1111/j.1471-8286.2005.01082.x
Vekemans X, Hardy O (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935. https://doi.org/10.1046/j.1365-294X.2004.02076.x
Vornam B, Decarli N, Gailing O (2004) Spatial distribution of genetic variation in a natural beech stand (Fagus sylvatica L.) based on microsatellite markers. Conserv Genet 5:561–570. https://doi.org/10.1023/B:COGE.0000041025.82917.ac
Vranckx G, Jacquemyn H, Mergeay J, Cox K, Kint V, Muys B, Honnay O (2014) Transmission of genetic variation from the adult generation to naturally established seedling cohorts in small forest stands of pedunculate oak (Quercus robur L.). For Ecol Manage 312:19–27. https://doi.org/10.1016/j.foreco.2013.10.027
Wagner AP, Creel S, Kalinowski ST (2006) Estimating relatedness and relationships using microsatellite loci with null alleles. Heredity 97:336–345. https://doi.org/10.1038/sj.hdy.6800865
Wang J (2005) Estimation of effective population sizes from data on genetic markers. Philos Trans R Soc B Biol Sci 360:1395–1409. https://doi.org/10.1098/rstb.2005.1682
Acknowledgements
This work was supported by the National Science Centre, Poland (Grant Numbers 2012/07/B/NZ9/00953). The authors wish to thank the referees for valuable and pertinent comments and Z. Kołodziej, PhD., for his help in collecting the field data.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Christian Ammer.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
OpenAccess This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Paluch, J., Zarek, M. & Kempf, M. The effect of population density on gene flow between adult trees and the seedling bank in Abies alba Mill.. Eur J Forest Res 138, 203–217 (2019). https://doi.org/10.1007/s10342-019-01162-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10342-019-01162-w