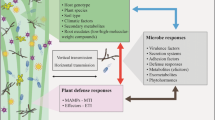
Abstract
Polyphagous aphids often have host-specialized lineages, and the mechanisms remain to be elucidated. The aphid Aphis gossypii has several host-specialized lineages including one specialized on Cucurbitaceae and one on Malvaceae. We found that the performance of Malvaceae lineage was poor on cucumbers, but significantly improved on cucumbers that were previously infested by Cucurbit lineage for 14 d. Following two-generation feeding experience on pre-infested cucumbers, Malvaceae lineage acquired the ability to use uninfested cucumbers. The pre-infestation largely decreased insect-negative metabolites such as cucurbitacins and phenols and increased insect-positive metabolites such as soluble sugars. The pre-infestation decreased the salicylic acid by 25.9% but increased the jasmonic acid by 1337% in cucumbers, which corresponded to expression of marker genes in phytohormone signaling pathways. Exogenous salicylic acid significantly decreased the performance improvement of Malvaceae lineage on pre-infested cucumbers, but exogenous jasmonic acid did not. Those results indicate that infestation by Cucurbit lineage altered the metabolism of cucumbers and interrupted the defense-associated phytohormones, from which we conclude that the disability in overcoming cucumber's defenses caused the incompatibility of Malvaceae lineage to cucumbers. The acclimation to cucumbers of Malvaceae lineage aphids may result from activation of some effector genes targeting cucumbers. Our findings provide new insights into the mechanisms of aphid host-specialization and new clues for preventing A. gossypii switching from Malvaceae hosts to cucurbits.






Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Aerts N, Pereira Mendes M, Van Wees SCM (2021) Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J 105:489–504. https://doi.org/10.1111/tpj.15124
Ali F, Hu X, Wang D et al (2021) Plant pathogen-mediated rapid acclimation of a host-specialized aphid to a non-host plant. Ecol Evol 11:15261–15272. https://doi.org/10.1002/ece3.8209
Bari R, Jones JDG (2009) Role of plant hormones in plant defence responses. Plant Mol Biol 69:473–488. https://doi.org/10.1007/s11103-008-9435-0
Belete T (2018) Defense mechanisms of plants to insect pests: from morphological to biochemical approach. Trends Tech Sci Res 2(2):30–38. https://doi.org/10.19080/ttsr.2018.02.555584
Blackman RL, Eastop VF (2007) Taxonomic issue. In: van Emden HF, Harrington R (eds) Aphids as crop pests. CAB International, London, pp 1–29
Boulain HF, Legeai J, Jaquiery E et al (2019) Differential expression of candidate salivary effector genes in pea aphid biotypes with distinct host plant specificity. Front Plant Sci 10:1301. https://doi.org/10.3389/fpls.2019.01301
Cao HH, Liu HR, Zhang ZF et al (2016) The green peach aphid Myzus persicae perform better on pre-infested Chinese cabbage Brassica pekinensis by enhancing host plant nutritional quality. Sci Rep 6:21954. https://doi.org/10.1038/srep21954
Chaudhary R, Atamian HS, Shen Z et al (2014) GroEL from the endosymbiont Buchnera aphidicola betrays the aphid by triggering plant defense. Proc Natl Acad Sci USA 111:8919–8924. https://doi.org/10.1073/pnas.1407687111
Chen Z, Zheng Z, Huang J et al (2009) Biosynthesis of salicylic acid in plants. Plant Signal Behav 4:493–496. https://doi.org/10.4161/psb.4.6.8392
Chen W, Gong L, Guo Z et al (2013) A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol Plant 6:1769–1780. https://doi.org/10.1093/mp/sst080
Chi H (2017) In TWOSEX-MSChart: a computer program for the age-stage, two-sex life table analysis. National Chung Hsing University, Taichung
Diezel C, von Dahl CC, Gaquerel E et al (2009) Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiol 150:1576–1586. https://doi.org/10.1104/pp.109.139550
Eleftherianos I, Vamvatsikos P, Ward D et al (2006) Changes in the levels of plant total phenols and free amino acids induced by two cereal aphids and effects on aphid fecundity. J Appl Entomol 130:15–19. https://doi.org/10.1111/j.1439-0418.2005.01017.x
Elzinga DA, De Vos M, Jander G (2014) Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol Plant Microbe Interact 27:747–756. https://doi.org/10.1094/MPMI-01-14-0018-R
Ferrater JB, Horgan FG (2016) Does Nilaparvata lugensgain tolerance to rice resistance genes through conspecifics at shared feeding sites? Entomol Exp Appl 160:77–82. https://doi.org/10.1111/eea.12454
Florencio-Ortiz NVO, Casas JL (2018) Local and systemic hormonal responses in pepper (Capsicum annuum L.) leaves under green peach aphid (Myzus persicae Sulzer) infestation. J Plant Physiol 231:356–363. https://doi.org/10.1016/j.jplph.2018.10.015
Gonzales WL, Ramırez CC, Olea N et al (2002) Host plant changes produced by the aphid Sipha flava: consequences for aphid feeding behaviour and growth. Entomol Exp Appl 103:107–113
Guldemond JA, Tigges WT, Vrijer PWF (1994) Host races of Aphis gossypii (Homoptera: Aphididae) on cucumber and chrysanthemum. Popul Ecol 23:1235–1240
Guo H, Yang F, Meng M et al (2022) No evidence of bacterial symbionts influencing host specificity in Aphis gossypii Glover (Hemiptera: Aphididae). InSects. https://doi.org/10.3390/insects13050462
Hogenhout SA, Bos JI (2011) Effector proteins that modulate plant-insect interactions. Curr Opin Plant Biol 14:422–428. https://doi.org/10.1016/j.pbi.2011.05.003
Kerchev PI, Fenton B, Foyer CH et al (2012) Plant responses to insect herbivory: interactions between photosynthesis, reactive oxygen species and hormonal signalling pathways. Plant Cell Environ 35:441–453. https://doi.org/10.1111/j.1365-3040.2011.02399.x
Koornneef A, Pieterse CM (2008) Cross talk in defense signaling. Plant Physiol 146:839–844. https://doi.org/10.1104/pp.107.112029
Koyama Y, Yao I, Akimoto SI (2004) Aphid galls accumulate high concentrations of amino acids: a support for the nutrition hypothesis for gall formation. Entomol Exp Appl 113:35–44
LeonardoTE MGT (2003) Facultative symbionts are associated with host plant specialization in pea aphid populations. Proc Biol Sci 270(Suppl 2):S209-212. https://doi.org/10.1098/rsbl.2003.0064
Liu XD, Zhai BP, Zhang XX et al (2002) Studies on the host biotypes and its cause of cotton aphid in Nanjing, China. Agric Sci China 1:1211–1215
Louis J, Shah J (2013) Arabidopsis thaliana-Myzus persicae interaction: shaping the understanding of plant defense against phloem-feeding aphids. Front Plant Sci 4:213. https://doi.org/10.3389/fpls.2013.00213
Mathers TC, Chen Y, Kaithakottil G et al (2017) Rapid transcriptional plasticity of duplicated gene clusters enables a clonally reproducing aphid to colonise diverse plant species. Genome Biol 18:27. https://doi.org/10.1186/s13059-016-1145-3
Mou DF, Kundu P, Pingault L et al (2023) Monocot crop–aphid interactions: plant resilience and aphid adaptation. Curr Opin Insect Sci 57:101038. https://doi.org/10.1016/j.cois.2023.101038
Najar-Rodríguez AJ, McGraw EA, Hull CD et al (2009) The ecological differentiation of asexual lineages of cotton aphids: alate behaviour, sensory physiology, and differential host associations. Biol J Linn Soc 97:503–519
Nguyen D, Rieu I, Mariani C et al (2016) How plants handle multiple stresses: hormonal interactions underlying responses to abiotic stress and insect herbivory. Plant Mol Biol 91:727–740. https://doi.org/10.1007/s11103-016-0481-8
Nomoto M, Skelly MJ, Itaya T et al (2021) Suppression of MYC transcription activators by the immune cofactor NPR1 fine-tunes plant immune responses. Cell Rep 37:110125. https://doi.org/10.1016/j.celrep.2021.110125
Nouhaud P, Peccoud J, Maheo F et al (2014) Genomic regions repeatedly involved in divergence among plant-specialized pea aphid biotypes. J Evol Biol 27:2013–2020. https://doi.org/10.1111/jeb.12441
O’Neal ME, Varenhorst AJ, Kaiser MC (2018) Rapid evolution to host plant resistance by an invasive herbivore: soybean aphid (Aphis glycines) virulence in North America to aphid resistant cultivars. Curr Opin Insect Sci 26:1–7. https://doi.org/10.1016/j.cois.2017.12.006
Ostrowski M, Jakubowska A (2014) Udp-glycosyltransferases of plant hormones. Adv Cell Biol 4:43–60. https://doi.org/10.2478/acb-2014-0003
Peccoud J, Simon JC, von Dohlen C et al (2010) Evolutionary history of aphid-plant associations and their role in aphid diversification. C R Biol 333:474–487. https://doi.org/10.1016/j.crvi.2010.03.004
Pitino M, Saskia AH (2013) Aphid protein effectors promote aphid colonization in a plant species-specific manner. MPMI 26:130–139. https://doi.org/10.1094/MPMI
Rodriguez PA, Escudero-Martinez C, Bos JI (2017) An aphid effector targets trafficking protein VPS52 in a host-specific manner to promote virulence. Plant Physiol 173:1892–1903. https://doi.org/10.1104/pp.16.01458
Sanchez-Arcos C, Reichelt M et al (2016) Modulation of legume defense signaling pathways by native and non-native pea aphid clones. Front Plant Sci. https://doi.org/10.3389/fpls.2016.01872
Shah J (2005) Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Ann Rev Phytopathol 43:229–260
Singh V, Shah J (2012) Tomato responds to green peach aphid infestation with the activation of trehalose metabolism and starch accumulation. Plant Signal Behav 7:605–607. https://doi.org/10.4161/psb.20066
Spoel SH, Johnson JS, Dong X (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA 104:18842–18847. https://doi.org/10.1073/pnas.0708139104
Storey JD, Leek JT, Bass AJ (2022) edge: extraction of differential gene expression. R package version 2.30.0. https://github.com/jdstorey/edge
Takemoto H, Uefune M, Ozawa R et al (2013) Previous infestation of pea aphids Acyrthosiphon pisumon broad bean plants resulted in the increased performance of conspecific nymphs on the plants. J Plant Interact 8:370–374. https://doi.org/10.1080/17429145.2013.786792
Tetyuk O, Benning UF, Hoffmann-Benning S (2013) Collection and analysis of Arabidopsis phloem exudates using the EDTA-facilitated method. J vis Exp. https://doi.org/10.3791/51111
Thaler JS, Humphrey PT, Whiteman NK (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci 17:260–270. https://doi.org/10.1016/j.tplants.2012.02.010
Tjallingii WF (1988) Electrical recording of stylet penetration activities. In: Minks AK, Harrewijn P (eds) Aphids, their biology, natural enemies and control. Elsevier Sci Publ, Amsterdam, pp 95–108
Tjallingii WF, Esch TH (1993) Fine structure of aphid stylet routes in plant tissues in correlation with EPG signals. Physiol Entomol 18:317–328
Tsuchida T, Koga R, Fukatsu T (2004) Host plant specialization governed by facultative symbiont. Science 303:1989. https://doi.org/10.1126/science.1094611
Underwood NC (1998) The timing of induced resistance and induced susceptibility in the soybean-Mexican bean beetle system. Oecologia 114:376–381
Varenhorst AJ, Michael T et al (2015) Determining the duration of Aphis glycines (Hemiptera: Aphididae) induced susceptibility effect in soybean. Arthropod Plant Interact 9:457–464. https://doi.org/10.1007/s11829-015-9395-7
Wang L, Zhang S, Luo JY et al (2016) Identification of Aphis gossypii Glover (Hemiptera: Aphididae) biotypes from different host plants in North China. PLoS ONE 11:e0146345. https://doi.org/10.1371/journal.pone.0146345
War AR, Sharma HC, Paulraj MG et al (2011) Herbivore induced plant volatiles: their role in plant defense for pest management. Plant Signal Behav 6:1973–1978. https://doi.org/10.4161/psb.6.12.18053
War AR, Paulraj MG, Ahmad T et al (2012) Mechanisms of plant defense against insect herbivores. Plant Signal Behav 7:1306–1320. https://doi.org/10.4161/psb.21663
Will T, Furch AC, Zimmermann MR (2013) How phloem-feeding insects face the challenge of phloem-located defenses. Front Plant Sci 4:336. https://doi.org/10.3389/fpls.2013.00336
Xia J, Guo Z, Yang Z et al (2021) Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 184(1693–1705):e1617. https://doi.org/10.1016/j.cell.2021.02.014
Xu HX, Qian LX, Wang XW et al (2019) A salivary effector enables whitefly to feed on host plants by eliciting salicylic acid-signaling pathway. Proc Natl Acad Sci USA 116:490–495. https://doi.org/10.1073/pnas.1714990116
Yang J, Duan G, Li C et al (2019) The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front Plant Sci 10:1349. https://doi.org/10.3389/fpls.2019.01349
Zarate SI, Kempema LA, Walling LL (2007) Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol 143:866–875. https://doi.org/10.1104/pp.106.090035
Zhang P, Zhu X, Huang F, Liu Y et al (2011) Suppression of jasmonic acid-dependent defense in cotton plant by the mealybug Phenacoccus solenopsis. PLoS ONE 6:e22378. https://doi.org/10.1371/journal.pone.0022378
Zhao H, Zhang X, Xue M et al (2015) Feeding of whitefly on tobacco decreases aphid performance via increased salicylate signaling. PLoS ONE 10:e0138584. https://doi.org/10.1371/journal.pone.0138584
Zhou SQ, Lou YR, Tzin V et al (2015) Alteration of plant primary metabolism in response to insect herbivory. Plant Physiol 169:1488–1498. https://doi.org/10.1104/pp.15.01405
Funding
This work was funded by the National Key Research and Development Project (2019YFD1002102), the National Natural Science Foundation of China (32171656) and STI 2030-Major Projects (2022ZD04021).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
This research did not involve human or higher animals.
Additional information
Communicated by Cesar Rodriguez-Saona.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hu, X., Hereward, J.P., Wang, D. et al. A host-specialized aphid lineage helps another conspecific lineage utilize a new host by disrupting the plant defenses. J Pest Sci (2023). https://doi.org/10.1007/s10340-023-01717-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10340-023-01717-2