Abstract
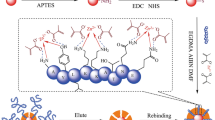
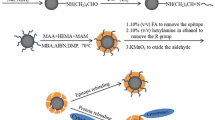
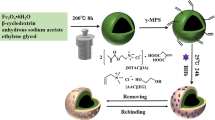
A kind of epitope surface imprinted particles was synthesized to selectively recognize cytochrome c (Cyt c) by a novel strategy assisted with γ-cyclodextrin (γ-CD) by host-guest interaction. C-terminal epitope nonapeptide of Cyt c was chosen as the template. γ-CD was immobilized on the surface of the silica as an encapsulated molecule to capture the template and improve the corresponding spatial orientation by the host-guest interaction for the peptide and target protein recognition. After γ-CD and epitope modified, the surface imprinted polymer assisted with the host-guest interaction was synthesized via monomers and cross-linkers by radical polymerization strategy. After the peptide removal, the epitope surface imprinted particles were obtained. The performance of Cyt c recognition in bovine serum sample by the imprinted polymers was calculated by HPLC. The modified imprinted polymers prepared achieved the best binding capability of 2.89 mg g−1 AE-9 with the IF = 4.07 and 37.58 mg g−1 Cyt c with the IF = 3.38, and it also realized an efficiency and selectivity protein recognition. Additionally, the imprinted particles assisting by γ-CD have good reusability with 89.89% of the original after five sorption-elution cycles and a better recognized ability (IF = 3.38) than those without γ-CD assisting (IF = 1.44). These results indicated that this epitope imprinted method assisted with the host-guest recognition interaction by γ-CD exhibited well specific recognition abilities towards the target protein Cyt c and potential application for Cyt c recognition in biological sample.
Graphical Abstract










Similar content being viewed by others
Data availability
The data that support the findings of this study is available in this publishing article of Chromatographia.
References
Chen L, Xu S, Li J (2011) Recent advances in molecular imprinting technology: current status, challenges and highlighted applications. Chem Soc Rev 40(5):2922. https://doi.org/10.1039/c0cs00084a
Hasanzadeh M, Shadjou N, Guardia M (2018) Cytosensing of cancer cells using antibody-based molecular imprinting: a short-review. Trends Anal Chem 99:129–134. https://doi.org/10.1016/j.trac.2017.12.010
Yang Q, Li J, Wang X et al (2018) Strategies of molecular imprinting-based fluorescence senors for chemical and biological analysis. Biosens Bioelectron 112:54–71. https://doi.org/10.1016/j.bios.2018.04.028
Tamayo FG, Turiel E, Martín-Esteban A (2007) Molecularly imprinted polymers for solid-phase extraction and solid-phase microextraction: recent developments and future trends. J Chromatogr A 1152(1–2):32–40. https://doi.org/10.1016/j.chroma.2006.08.095
Yola ML, Eren T, Atar N (2014) Molecularly imprinted electrochemical biosensor based on Fe@Au nanoparticles involved in 2-aminoethanethiol functionalized multi-walled carbon nanotubes for sensitive determination of cefexime in human plasma. Biosens Bioelectron 60:277–285. https://doi.org/10.1016/j.bios.2014.04.045
Tada M, Sasaki T, Iwasawa Y (2004) Design of a novel molecular-imprinted Rh-amine complex on SiO2 and its shape-selective catalysis for α-methylstyrene hydrogenation. J Phys Chem B 108(9):2918–2930. https://doi.org/10.1021/jp036421v
Kempe M, Mosbach K (1995) Separation of amino acids, peptides and proteins on molecularly imprinted stationary phases. J Chromatogr A 691(1–2):317–323. https://doi.org/10.1016/0021-9673(94)00820-y
Ye L (2015) Synthetic strategies in molecular imprinting. Adv Biochem Eng/Biotechnol. https://doi.org/10.1007/10_2015_313
Hua Z, Zhou S, Zhao M (2009) Fabrication of a surface imprinted hydrogel shell over silica microspheres using bovine serum albumin as a model protein template. Biosens Bioelectron 25(3):615–622. https://doi.org/10.1016/j.bios.2009.01.027
Lu Y, Li CX, Liu XH et al (2009) Molecular recognition through the exact placement of functional groups on non-covalent molecularly imprinted polymers. J Chromatogr A 950(1–2):89–97. https://doi.org/10.1016/s0021-9673(02)00058-4
Luo J, Huang J, Cong J et al (2009) Double recognition and selective extraction of glycoprotein based on the molecular imprinted graphene oxide and boronate affinity. ACS Appl Mater Interfaces 9(8):7735–7744. https://doi.org/10.1021/acsami.6b14733
Li SJ, Cao SS, Whitcombe MJ et al (2014) Size matters: challenges in imprinting macromolecules. Prog Polym Sci 39(1):145–163. https://doi.org/10.1016/j.progpolymsci.2013.10.0
Ge Y, Turner APF (2008) Too large to fit? Recent developments in macromolecular imprinting. Trends Biotechnol 26(4):218–224. https://doi.org/10.1016/j.tibtech.2008.01.001
Li SW, Yang KG, Liu JX et al (2015) Surface-imprinted nanoparticles prepared with a his-tag-anchored epitope as the template. Anal Chem 87(9):4617–4620. https://doi.org/10.1021/ac5047246
Wu G, Li JY, Qu X et al (2015) Template size matched film thickness for effectively in situ surface imprinting: a model study of glycoprotein imprints. RSC Adv 5(58):47010–47021. https://doi.org/10.1039/c5ra06454f
Yang KG, Zhang LH, Liang Z et al (2012) Protein-imprinted materials: rational design, application and challenges. Anal Bioanal Chem 403(8):2173–2183. https://doi.org/10.1007/s00216-012-5840-y
Yang X, Sun Y, Xiang Y et al (2019) Controlled synthesis of PEGylated surface protein-imprinted nanoparticles. Analyst 144(18):5439–5448. https://doi.org/10.1039/c9an01221d
Li Q, Yang K, Li S et al (2015) Preparation of surface imprinted core-shell particles via a metal chelating strategy: specific recognition of porcine serum albumin. Microchim Acta 183(1):345–352. https://doi.org/10.1007/s00604-015-1640-3
Zhang XM, Qin YP, Ye HL et al (2018) Silicon nanoparticles coated with an epitope-imprinted polymer for fluorometric determination of cytochrome c. Mikrochim Acta 185(3):173–182. https://doi.org/10.1007/s00604-018-2724-7
Gao R, Mu X, Hao Y et al (2014) Combination of surface imprinting and immobilized template techniques for preparation of core-shell molecularly imprinted polymers based on directly amino-modified Fe3O4 nanoparticles for specific recognition of bovine hemoglobin. J Mater Chem B 22(12):1733–1741. https://doi.org/10.1039/c3tb21684e
Rachkov A, Minoura N (2000) Recognition of oxytocin and oxytocin-related peptides in aqueous media using a molecularly imprinted polymer synthesized by the epitope approach. J Chromatogr A 889(1–2):111–118. https://doi.org/10.1016/s0021-9673(00)00568-9
Kryscio DR, Peppas NA (2012) Critical review and perspective of macromolecularly imprinted polymers. Acta Biomater 8(2):461–473. https://doi.org/10.1016/j.actbio.2011.11.005
Sette A, Fikes J (2003) Epitope-based vaccines: an update on epitope identification, vaccine design and delivery. Curr Opin Immunol 15(4):461–470. https://doi.org/10.1016/s0952-7915(03)00083-9
Rachkov A, Minoura N (2001) Towards molecularly imprinted polymers selective to peptides and proteins. The epitope approach. Biochimica Et Biophysica Acta-Protein Struct Mol Enzymol 1544(1–2):255–266. https://doi.org/10.1016/s0167-4838(00)00226-0
Reichlin M (1972) Localizing antigenic determinants in human haemoglobin with mutants: molecular correlations of immunological tolerance. J Mol Biol 64(2):485–496. https://doi.org/10.1016/0022-2836(72)90512-8
Nishino H, Huang CS, Shea KJ (2006) Selective protein capture by epitope imprinting. Angewandte Chemie-International Edition 45(15):2392–2396. https://doi.org/10.1002/anie.200503760
Moczko E, Guerreiro A, Caceres C et al (2019) Epitope approach in molecular imprinting of antibodies. J Chromatogr B-Anal Technol Biomed Life Sci 124:1–6. https://doi.org/10.1016/j.jchromb.2019.05.024
Yang YQ, He XW, Wang YZ et al (2014) Epitope imprinted polymer coating CdTe quantum dots for specific recognition and direct fluorescent quantification of the target protein bovine serum albumin. Biosens Bioelectron 54:266–272. https://doi.org/10.1016/j.bios.2013.11.004
Yang KG, Liu JX, Li SW et al (2014) Epitope imprinted polyethersulfone beads by self-assembly for target protein capture from the plasma proteome. Chem Commun 50(67):9521–9524. https://doi.org/10.1039/c4cc03428g
Li Z, Guan P, Hu X, Ding S, Tian Y, Xu Y, Qian L (2018) Preparation of molecularly imprinted mesoporous materials for highly enhancing adsorption performance of cytochrome C. Polymers 10(3):298–311. https://doi.org/10.3390/polym10030298
Zhang W, Zhang T, Chen Y (2018) Simultaneous quantification of Cyt c interactions with HSP27 and Bcl-xL using molecularly imprinted polymers (MIPs) coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based targeted proteomics. J Proteomics 192:188–195. https://doi.org/10.1016/j.jprot.2018.09.001
Whitcombe MJ, Chianella I, Larcombe L et al (2011) The rational development of molecularly imprinted polymer-based sensors for protein detection. Chem Soc Rev 40(3):1547–1571. https://doi.org/10.1039/c0cs00049c
D’Aria F, Serri C, Niccoli M, Mayol L, Quagliariello V, Iaffaioli RV, Giancola C (2017) Host–guest inclusion complex of quercetin and hydroxypropyl-β-cyclodextrin. J Therm Anal Calorim 130(1):451–456. https://doi.org/10.1007/s10973-017-6135-5
Zhang W, Qin L, He XW, Li WY, Zhang YK (2009) Novel surface modified molecularly imprinted polymer using acryloyl-β-cyclodextrin and acrylamide as monomers for selective recognition of lysozyme in aqueous solution. J Chromatogr A 1216(21):4560–4567. https://doi.org/10.1016/j.chroma.2009.03.056
Song S, Shirasaka K, Katayama M, Nagaoka S, Yoshihara S, Osawa T, Komiyama M (2007) Recognition of solution structures of peptides by molecularly imprinted cyclodextrin polymers. Macromolecules 40(10):3530–3532. https://doi.org/10.1021/ma070348f
Min Y, Jiang B, Wu C et al (2014) 1.9 mum superficially porous packing material with radially oriented pores and tailored pore size for ultra-fast separation of small molecules and biomolecules. J Chromatogr A 1356:148–156. https://doi.org/10.1016/j.chroma.2014.06.049
Qin L, He XW, Li WY, Zhang YK (2008) Molecularly imprinted polymer prepared with bonded β-cyclodextrin and acrylamide on functionalized silica gel for selective recognition of tryptophan in aqueous media. J Chromatogr A 1187(1–2):94–102. https://doi.org/10.1016/j.chroma.2008.02.004
Qin YP, Wang HY, He XW, Li WY, Zhang YK (2018) Metal chelation dual-template epitope imprinting polymer via distillation-precipitation polymerization for recognition of porcine serum albumin. Talanta 185:620–627. https://doi.org/10.1016/j.talanta.2018.03.082
Acknowledgements
This work was supported by National Natural Science Foundation of China (No. 21804099) and Open Project of Tianjin Key Laboratory of Biomedical Materials (No. 2022BMEKFKT003).
Funding
This work was funded by National Natural Science Foundation of China (Grant Number 21804099) and Open Project of Tianjin Key Laboratory of Biomedical Materials (Grant Number 2022BMEKFKT003). Qinran Li and Hongfeng Zhang declared that the funds were supported by National Natural Science Foundation of China (Grant Number 21804099). Wenquan Ji, Hongfeng Zhang, Jin Zhao, Qinran Li and Donglan Sun declared that the funds were supported by Open Project of Tianjin Key Laboratory of Biomedical Materials (Grant Number 2022BMEKFKT003). Yongjian Wang, Qiliang Deng and Tianjun Liu declared that there was no fund support in the research process. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Wenquan Ji and Yongjian Wang proceed the main experiment and wrote the main manuscript text. Hongfeng Zhang established chromatographic separation methods and provided support for the use of chromatographic instruments. Jin Zhao provided the support for functional modification of γ-cyclodextrin and peptide inclusion. Qinran Li revised the whole manuscript and finished the final draft. Qiliang Deng, Donglan Sun and Tianjun Liu put forward some useful suggestions for the revision and improvement of the manuscript. All of the authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ji, W., Wang, Y., Zhang, H. et al. Preparation of C-Terminal Epitope Imprinted Particles for Recognition of Cytochrome c Assisted with γ-Cyclodextrin by Host-Guest Interaction. Chromatographia 86, 469–482 (2023). https://doi.org/10.1007/s10337-023-04257-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10337-023-04257-0