Abstract
Objective
To design, develop and evaluate an automated multi-atlas method for segmentation and volume quantification of gluteus maximus from Dixon and T1-weighted images.
Materials and methods
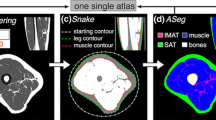
The multi-atlas segmentation method uses an atlas library constructed from 15 Dixon MRI scans of healthy subjects. A non-rigid registration between each atlas and the target, followed by majority voting label fusion, is used in the segmentation. We propose a region of interest (ROI) to standardize the measurement of muscle bulk. The method was evaluated using the dice similarity coefficient (DSC) and the relative volume difference (RVD) as metrics, for Dixon and T1-weighted target images.
Results
The mean(± SD) DSC was 0.94 ± 0.01 for Dixon images, while 0.93 ± 0.02 for T1-weighted. The RVD between the automated and manual segmentation had a mean(± SD) value of 1.5 ± 4.3% for Dixon and 1.5 ± 4.8% for T1-weighted images. In the muscle bulk ROI, the DSC was 0.95 ± 0.01 and the RVD was 0.6 ± 3.8%.
Conclusion
The method allows an accurate fully automated segmentation of gluteus maximus for Dixon and T1-weighted images and provides a relatively accurate volume measurement in shorter times (~ 20 min) than the current gold-standard manual segmentations (2 h). Visual inspection of the segmentation would be required when higher accuracy is needed.







Similar content being viewed by others
References
Anderson FC, Pandy MG (2003) Individual muscle contributions to support in normal walking. Gait Posture 17:159–169
Bartlett JL, Sumner B, Ellis RG, Kram R (2014) Activity and functions of the human gluteal muscles in walking, running, sprinting, and climbing. Am J Phys Anthropol 153:124–131
Lieberman DE, Raichlen DA, Pontzer H, Bramble DM, Cutright-Smith E (2006) The human gluteus maximus and its role in running. J Exp Biol 209:444–454
Ward SR, Winters TM, Blemker SS (2010) The architectural design of the gluteal muscle group: implications for movement and rehabilitation. J Orthop Sport Phys Ther 40:95–102
Zacharias A, Pizzari T, English DJ, Kapakoulakis T, Green RA (2016) Hip abductor muscle volume in hip osteoarthritis and matched controls. Osteoarthr Cartil 24:1727–1735
Grimaldi A, Richardson C, Stanton W, Durbridge G, Donnelly W, Hides J (2009) The association between degenerative hip joint pathology and size of the gluteus medius, gluteus minimus and piriformis muscles. Man Ther 14:605–610
Uemura K, Takao M, Sakai T, Nishii T, Sugano N (2016) Volume increases of the gluteus maximus, gluteus medius, and thigh muscles after hip arthroplasty. J Arthroplasty 31:906–912.e1
Smith MMF, Bonacci J, Mendis MD, Christie C, Rotstein A, Hides JA (2017) Gluteus medius activation during running is a risk factor for season hamstring injuries in elite footballers. J Sci Med Sport 20:159–163
Sakamaki M, Bemben MG, Abe T (2011) Legs and trunk muscle hypertrophy following walk training with restricted leg muscle blood flow. J Sports Sci Med 10:338–340
Mendieta CG, Sood A (2018) Classification system for gluteal evaluation revisited. Clin Plast Surg. https://doi.org/10.1016/j.cps.2017.12.013
O’Brien TD, Reeves ND, Baltzopoulos V, Jones DA, Maganaris CN (2009) Strong relationships exist between muscle volume, joint power and whole-body external mechanical power in adults and children. Exp Physiol 94:731–738
Akagi R, Suzuki M, Kawaguchi E, Miyamoto N, Yamada Y, Ema R (2018) Muscle size-strength relationship including ultrasonographic echo intensity and voluntary activation level of a muscle group. Arch Gerontol Geriatr 75:185–190
Dawes H, Smith C, Collett J, Wade D, Howells K, Ramsbottom R, Izadi H, Sackley C (2005) A pilot study to investigate explosive leg extensor power and walking performance after stroke. J Sports Sci Med 4:556–562
Bassey EJ, Fiatarone MA, O’Neill EF, Kelly M, Evans WJ, Lipsitz LA (1992) Leg extensor power and functional performance in very old men and women. Clin Sci (Lond) 82:321–327
Cronin J, Sleivert G (2005) Challenges in understanding the influence of maximal power training on improving athletic performance. Sport Med 35:213–234
Bley TA, Wieben O, François CJ, Brittain JH, Reeder SB (2010) Fat and water magnetic resonance imaging. J Magn Reson Imaging 31:4–18
Xiang QS, An L (1997) Water-fat imaging with direct phase encoding. J Magn Reson Imaging 7:1002–1015
Hernando D, Liang Z-P, Kellman P (2010) Chemical shift-based water/fat separation: a comparison of signal models. Magn Reson Med 64:811–822
Dixon WT (1984) Simple proton spectroscopic imaging. Radiology 153:189–194
Wokke BH, Bos C, Reijnierse M, Van Rijswijk CS, Eggers H, Webb A, Verschuuren JJ, Kan HE (2013) Comparison of dixon and T1-weighted MR methods to assess the degree of fat infiltration in duchenne muscular dystrophy patients. J Magn Reson Imaging 38:619–624
Barnouin Y, Butler-Browne G, Voit T, Reversat D, Azzabou N, Leroux G, Behin A, McPhee JS, Carlier PG, Hogrel JY (2014) Manual segmentation of individual muscles of the quadriceps femoris using MRI: a reappraisal. J Magn Reson Imaging 40:239–247
Karlsson A, Rosander J, Romu T, Tallberg J, Grönqvist A, Borga M, Dahlqvist Leinhard O (2015) Automatic and quantitative assessment of regional muscle volume by multi-atlas segmentation using whole-body water-fat MRI. J Magn Reson Imaging 41:1558–1569
Kovanlikaya A, Mittelman SD, Ward A, Geffner ME, Dorey F, Gilsanz V (2005) Obesity and fat quantification in lean tissues using three-point Dixon MR imaging. Pediatr Radiol 35:601–607
Belavý DL, Miokovic T, Rittweger J, Felsenberg D (2011) Estimation of changes in volume of individual lower-limb muscles using magnetic resonance imaging (during bed-rest). Physiol Meas 32:35–50
Nordez A, Jolivet E, Südhoff I, Bonneau D, De Guise JA, Skalli W (2009) Comparison of methods to assess quadriceps muscle volume using magnetic resonance imaging. J Magn Reson Imaging 30:1116–1123
Voronov AV (2003) Anatomical cross-sectional areas and volumes of the muscles of the lower extremities. Hum Physiol 29:201–211
Mersmann F, Bohm S, Schroll A, Arampatzis A (2014) Validation of a simplified method for muscle volume assessment. J Biomech 47:1348–1352
Pedoia V, Majumdar S, Link TM (2016) Segmentation of joint and musculoskeletal tissue in the study of arthritis. Magn Reson Mater Phy 29:207–221
Baudin PY, Azzabou N, Carlier PG, Paragios N (2012) Automatic skeletal muscle segmentation through random walks and graph-based seed placement. In: Proceedings—international symposium on biomedical imaging. IEEE, pp 1036–1039
Orgiu S, Lafortuna CL, Rastelli F, Cadioli M, Falini A, Rizzo G (2016) Automatic muscle and fat segmentation in the thigh from T 1-weighted MRI. J Magn Reson Imaging 43:601–610
Brunner G, Nambi V, Yang E, Kumar A, Virani SS, Kougias P, Shah D, Lumsden A, Ballantyne CM, Morrisett JD (2011) Automatic quantification of muscle volumes in magnetic resonance imaging scans of the lower extremities. Magn Reson Imaging. https://doi.org/10.1016/j.mri.2011.02.033
Heckemann RA, Hajnal JV, Aljabar P, Rueckert D, Hammers A (2006) Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. Neuroimage 33:115–126
Iglesias JE, Sabuncu MR (2015) Multi-atlas segmentation of biomedical images: a survey. Med Image Anal 24:205–219
Le Troter A, Fouré A, Guye M, Confort-Gouny S, Mattei JP, Gondin J, Salort-Campana E, Bendahan D (2016) Volume measurements of individual muscles in human quadriceps femoris using atlas-based segmentation approaches. Magn Reson Mater Phy 29:245–257
Scheys L, Loeckx D, Spaepen A, Suetens P, Jonkers I (2009) Atlas-based non-rigid image registration to automatically define line-of-action muscle models: a validation study. J Biomech 42:565–572
Ghosh S, Ray N, Boulanger P (2017) A structured deep-learning based approach for the automated segmentation of human leg muscle from 3D MRI. In: 2017 14th Conf. Comput. Robot Vis. IEEE, pp 117–123
Ogier A, Sdika M, Foure A, Le Troter A, Bendahan D (2017) Individual muscle segmentation in MR images: a 3D propagation through 2D non-linear registration approaches. In: Proceedings of annual international conference of the IEEE engineering in medicine and biology society. EMBS. IEEE, pp 317–320
Ranzini MBM, Ebner M, Cardoso MJ, Fotiadou A, Vercauteren T, Henckel J, Hart A, Ourselin S, Modat M (2018) Joint multimodal segmentation of clinical CT and MR from hip arthroplasty patients. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics). Springer, Cham, pp 72–84
Klemt C, Modat M, Pichat J, Cardoso MJ, Henckel J, Hart A, Ourselin S (2015) Automatic assessment of volume asymmetries applied to hip abductor muscles in patients with hip arthroplasty. Proc. SPIE 9413, Medical Imaging 2015: Image Processing, 94131M
Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29:1310–1320
Rueckert D, Sonoda LI, Hayes C, Hill DLG, Leach MO, Hawkes DJ (1999) Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging 18:712–721
Marstal K, Berendsen F, Staring M, Klein S (2016) SimpleElastix: a user-friendly, multi-lingual library for medical image registration. In: 2016 IEEE conference on computer vision and pattern recognition work. IEEE, pp 574–582
Klein S, Staring M, Murphy K, Viergever MA, Pluim JPW (2010) elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging. https://doi.org/10.1109/TMI.2009.2035616
Avants BB, Yushkevich P, Pluta J, Minkoff D, Korczykowski M, Detre J, Gee JC (2010) The optimal template effect in hippocampus studies of diseased populations. Neuroimage 49:2457–2466
Klein S, Pluim JPW, Staring M, Viergever MA (2009) Adaptive stochastic gradient descent optimisation for image registration. Int J Comput Vis 81:227–239
Aljabar P, Heckemann RA, Hammers A, Hajnal JV, Rueckert D (2009) Multi-atlas based segmentation of brain images: atlas selection and its effect on accuracy. Neuroimage 46:726–738
Xu L, Krzyzak A, Suen CY (1992) Methods of combining multiple classifiers and their applications to handwriting recognition. IEEE Trans Syst Man Cybern 22:418–435
Kolk S, Klawer EME, Schepers J, Weerdesteyn V, Visser EP, Verdonschot N (2015) Muscle activity during walking measured using 3D MRI segmentations and [18F]-fluorodeoxyglucose in combination with positron emission tomography. Med Sci Sports Exerc 47:1896–1905
Addison O, Marcus RL, Lastayo PC, Ryan AS (2014) Intermuscular fat: a review of the consequences and causes. Int J Endocrinol. https://doi.org/10.1155/2014/309570
Warfield SK, Zou KH, Wells WM (2004) Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 23:903–921
Jorge Cardoso M, Leung K, Modat M, Keihaninejad S, Cash D, Barnes J, Fox NC, Ourselin S (2013) STEPS: similarity and truth estimation for propagated segmentations and its application to hippocampal segmentation and brain parcelation. Med Image Anal 17:671–684
Bojorquez JZ, Bricq S, Walker PM, Lalande A (2015) Automatic classification of tissues using T1 and T2 relaxation times from prostate MRI: a step towards generation of PET/MR attenuation map. In: 2015 IEEE international conference on image processing. IEEE, pp 1185–1189
Yang YX, Chong MS, Tay L, Yew S, Yeo A, Tan CH (2016) Automated assessment of thigh composition using machine learning for Dixon magnetic resonance images. Magn Reson Mater Phy 29:723–731
Wang G, Li W, Ourselin S, Vercauteren T (2018) Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks. In: Crimi A, Bakas S, Kuijf H, Menze B, Reyes M (eds) Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. BrainLes 2017. Lecture notes in computer science, vol 10670. Springer, Cham
Perone CS, Calabrese E, Cohen-Adad J (2018) Spinal cord gray matter segmentation using deep dilated convolutions. Sci Rep 8:5966
Zhou Z, Zhao G, Kijowski R, Liu F (2018) Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med 80:2759–2770
Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R (2018) Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med 79:2379–2391
Acknowledgements
This research study was funded by The Maurice Hatter Foundation, the RNOH Charity, the Rosetrees Trust and the Stoneygate Trust and supported by researchers at the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Funding
This research study was funded by The Maurice Hatter Foundation, the RNOH Charity, the Rosetrees Trust and the Stoneygate Trust and supported by researchers at the National Institute for Health Research University College London Hospitals Biomedical Research Centre.
Author information
Authors and Affiliations
Contributions
MAB: Study conception and design, acquisition of data, analysis and interpretation of data, drafting of manuscript, critical revision. JH: Study conception and design, acquisition of data, analysis and interpretation of data, critical revision. AF: Acquisition of data, critical revision. ADL: Study conception and design, Acquisition of data, critical revision. AH: Study conception and design, acquisition of data, analysis and interpretation of data, critical revision.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in this work involving human participants were approved by the local Institutional Review Board. All subjects have given written informed consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Belzunce, M.A., Henckel, J., Fotiadou, A. et al. Automated multi-atlas segmentation of gluteus maximus from Dixon and T1-weighted magnetic resonance images. Magn Reson Mater Phy 33, 677–688 (2020). https://doi.org/10.1007/s10334-020-00839-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10334-020-00839-3